- The paper introduces an automated framework that uses segmentation-based metrics to quantify staining accuracy in virtual IHC images.
- The paper benchmarks 16 deep learning models across paired and unpaired paradigms, revealing discrepancies between conventional fidelity metrics and biologically accurate segmentation outcomes.
- The paper highlights challenges in whole slide image evaluation and emphasizes scalable, context-aware assessments for clinical translation.
Building Trust in Virtual Immunohistochemistry: Automated Assessment of Image Quality
Motivation and Problem Context
The synthesis of virtual immunohistochemistry (IHC) stains from hematoxylin and eosin (H&E) images using deep learning offers significant clinical value by reducing the cost and time of molecular diagnostics in pathology. However, the reliability of such virtual stains is constrained by the limitations of conventional image quality assessment metrics, which have not been systematically validated to capture the accuracy of IHC-positive cell labeling. This work presents an automated framework for evaluating virtual IHC quality—across both paired and unpaired image translation models—by introducing segmentation-based metrics that directly quantify the staining correctness at pixel-level, independent of expert annotation.
Comprehensive Evaluation Framework
The approach encompasses both fidelity and stain accuracy assessments, providing a multi-criteria evaluation benchmark for generated IHC images. The workflow integrates paired and unpaired model training paradigms, leveraging both registered tiles and whole-slide images (WSIs), and computes a suite of metrics, ranging from established distribution and texture scores (e.g., FID, KID, PSNR, SSIM) to segmentation-based accuracy indices (DICE, IoU, Hausdorff distance, TPR, TNR).
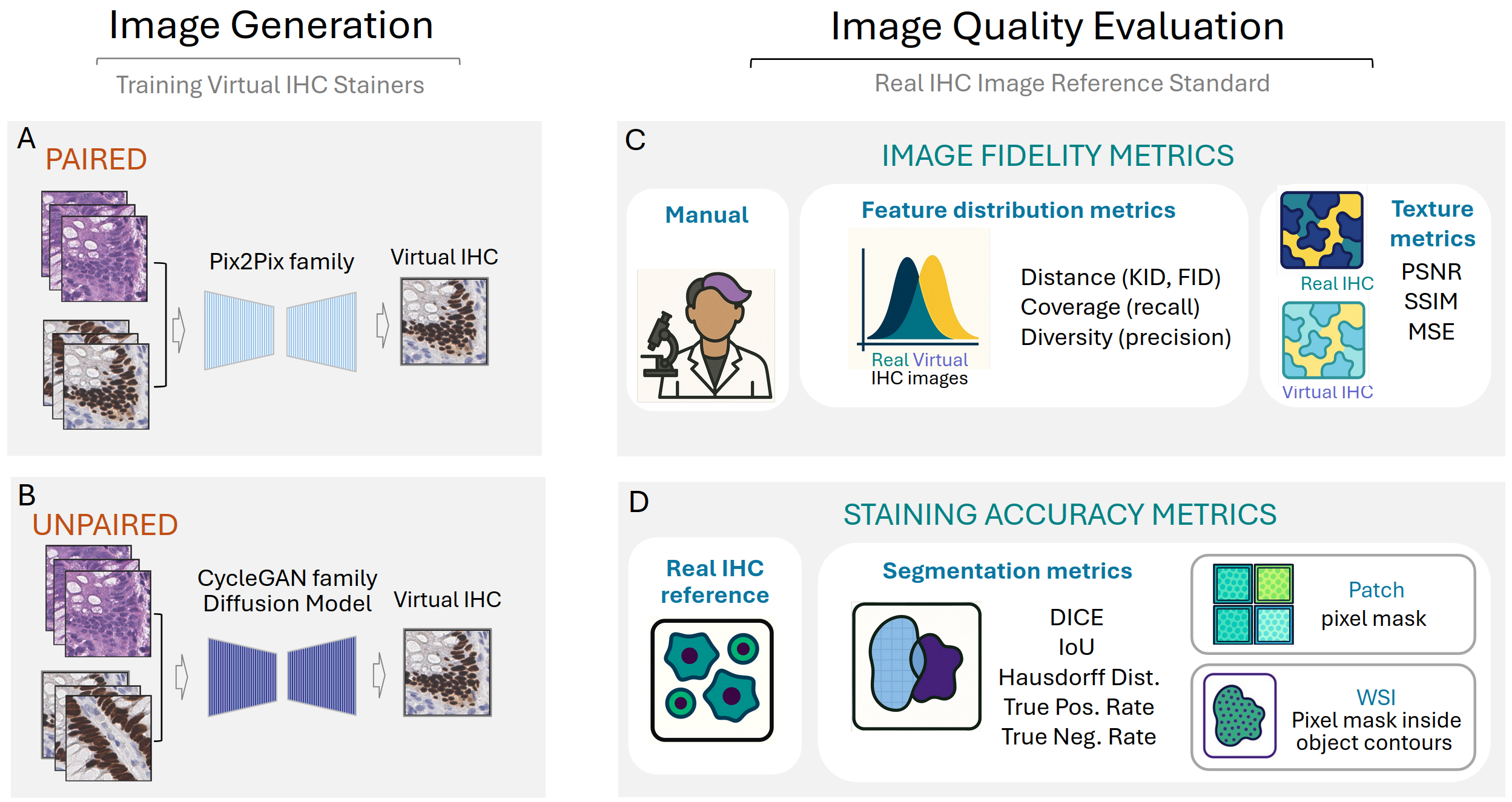
Figure 1: Workflow for virtual IHC image generation and multi-axis evaluation, outlining the distinction between paired/unpaired model training, chosen quality metrics, and accuracy scoring protocols for tiles and WSIs.
Model Architectures and Training Paradigms
Sixteen image-to-image translation models are benchmarked, spanning paired models (Pix2Pix family, including PyramidPix2Pix and AdaptiveNCE) and unpaired models (CycleGAN family, recent variants, and diffusion-based architectures like UNSB). Paired approaches utilize pixel-aligned H&E—IHC training data, while unpaired methods learn from non-aligned samples, exploiting cycle-consistency or adversarial and diffusion objectives to generate realistic stains.
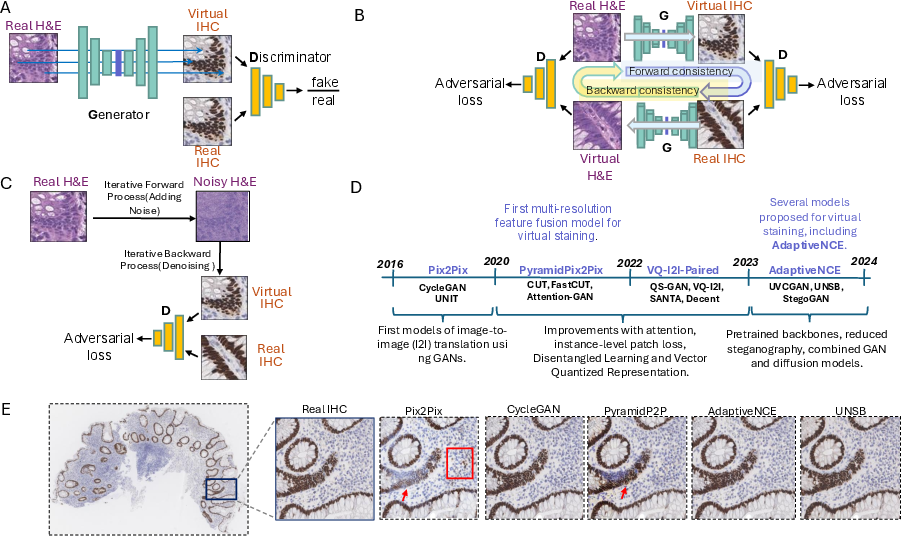
Figure 2: Overview of architectural families, including training mechanisms for Pix2Pix (paired, adversarial), CycleGAN (unpaired, cycle-consistent), and diffusion-based models; timeline contextualizes evolution in methodological rigor and performance.
Fidelity Metrics: Limitations and Statistical Correlations
Evaluation using FID, KID, feature distribution precision/recall, PSNR, and SSIM reveals that unpaired models frequently yield superior distribution metrics compared to paired models, except for AdaptiveNCE which matches SANTA in FID/KID due to architectural enhancements. Notably, manual pathologist assessment of virtual image quality yields results poorly correlated with FID scores (r=−0.034) and only moderately aligned at the model level with PSNR, SSIM, and MSE, yet providing little reliability at the image patch level.
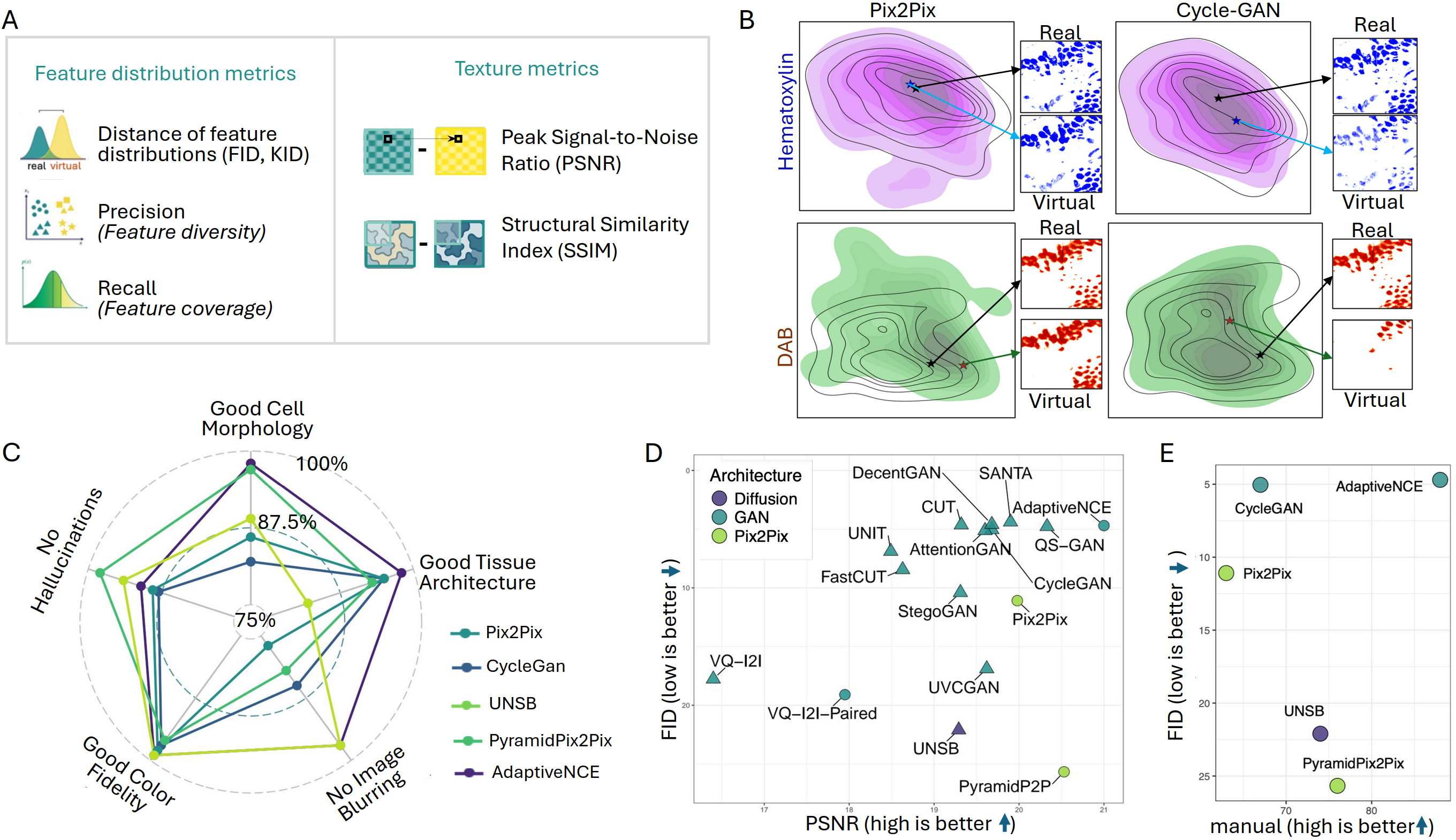
Figure 3: Quantitative and qualitative assessment modalities: distribution/texture metrics, UMAP-based feature overlap for blue and brown stain channels, and manual scoring for cell/tissue architecture.
Segmentation-Based Stain Accuracy: Interpretability and Robustness
Segmentation-based metrics—derived by decomposing IHC images into DAB masks (via color deconvolution or trained U-Net segmentation)—provide direct, scale-adaptive quantification of positive cell labeling accuracy. Paired models achieve statistically superior DICE and IoU compared to unpaired, but true positive rates remain suboptimal for all models, indicating challenges in accurate IHC-positive detection at the pixel level.
The correlation between fidelity and accuracy metrics is negligible for FID vs. DICE (r=0.002), and patch-level relationships between texture and accuracy metrics are inconsistent or negatively correlated, underscoring orthogonality between perceptual image similarity and biological signal preservation.
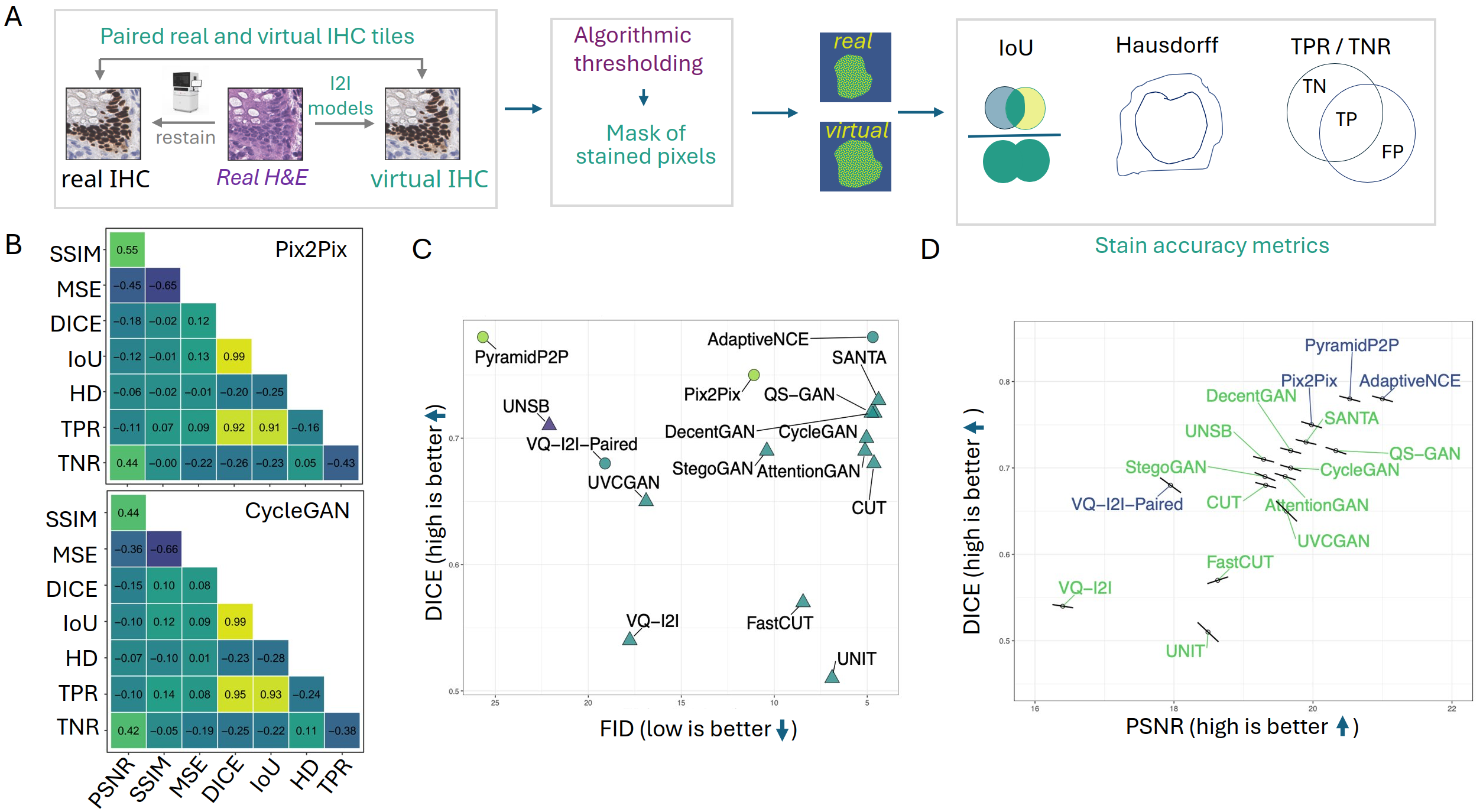
Figure 4: Segmentation workflow for DAB mask generation and metric computation; correlation analyses emphasize the independence of fidelity and accuracy measures at both model and tile levels.
While tile-based assessments are standard, WSIs introduce new challenges regarding spatial continuity, artifact propagation at tile boundaries, and decline in segmentation accuracy due to overlap-based smoothing. Even top models exhibit substantial metric degradation in WSI gland segmentation outputs compared to their patch-level performance, confirming the critical importance of large-scale, context-aware evaluation for clinical translation.
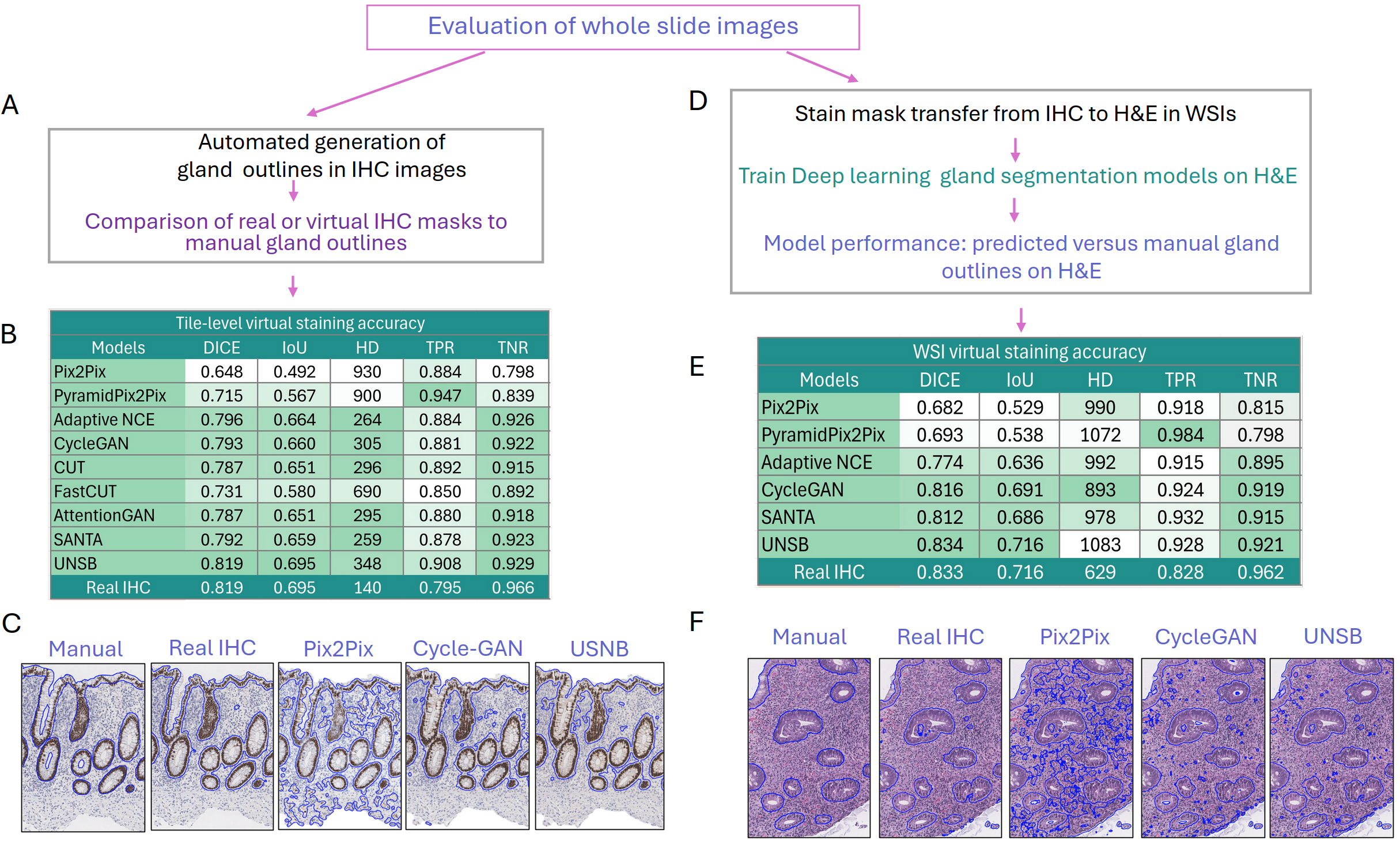
Figure 5: WSI-level gland segmentation performance and qualitative comparisons versus manual annotation; block-like tiling artifacts and loss of discriminative detail are highlighted, particularly in paired models.
Implementation and Scaling Considerations
All models are retrained on a unified dataset of CDX2 restained biopsies, with preprocessing pipeline steps for tissue extraction, registration, and balanced patch sampling to ensure robust, unbiased evaluation. GPU resource utilization is dominated by diffusion-based and multi-scale architectures, and all process modules are available in open-source repositories to maximize reproducibility. Segmentation thresholds, U-Net model settings, and tile aggregation protocols are key hyperparameters influencing accuracy metrics in deployment.
Implications and Future Directions
The paper decisively confirms that conventional distribution/texture metrics are not reliable surrogates for biologically meaningful stain accuracy in virtual IHC, challenging the methodological status quo in generative image evaluation for medical domains. Automated, pathologist-free assessments using paired, pixel-level ground truth and segmentation-based metrics offer the only scalable, interpretable route for benchmarking and clinical validation.
In future research, the objective is to adapt accuracy-based protocols to unpaired datasets, develop models with improved global spatial consistency for WSIs, and extend methodology to additional stain modalities (autofluorescence, unstained-to-stained synthesis) where manual annotation is inherently unreliable. Comprehensive metric frameworks must be tailored to downstream clinical application—prediction, diagnosis, quantification—and not treated as agnostic measures of generative model success.
Conclusion
Automated segmentation-based evaluation protocols are indispensable for establishing trust in virtual IHC images produced by deep generative models. This work provides clear evidence that texture and distribution metrics alone cannot guarantee accurate biological signal representation, especially in critical diagnostic contexts. For translation into clinical workflows, future model architectures must optimize both visual fidelity and biological stain accuracy, and all evaluations must be made context- and task-specific, ideally incorporating WSI-scale assessments. The methodology, code, and data resources presented herein will facilitate benchmarking, comparison, and acceleration of reliable virtual staining technologies for pathology.




