- The paper introduces Curia, a multi-modal radiology model trained on 200M CT/MRI images using a DINOv2-driven self-supervised Vision Transformer approach.
- It demonstrates high data efficiency and robust cross-modality generalization, outperforming existing models and achieving near-perfect accuracy in anatomical classification.
- Curia delivers strong performance across 19 benchmark tasks including classification, segmentation, registration, oncology, and survival prediction, matching or exceeding expert radiologists.
Curia: A Multi-Modal Foundation Model for Radiology
Introduction and Motivation
Curia is introduced as a multi-modal radiological foundation model (FM) trained on an unprecedented scale: 200 million CT and MRI images (130 TB) from 150,000 clinical exams. The model leverages self-supervised learning via the DINOv2 algorithm and is built on the Vision Transformer (ViT) architecture. The primary motivation is to overcome the limitations of narrow, single-task radiological models, which require extensive manual annotation and lack generalizability across modalities and clinical tasks. Curia aims to provide broad generalization, strong performance in low-data regimes, and emergent cross-modal capabilities, evaluated on a comprehensive 19-task benchmark (CuriaBench) spanning classification, regression, survival prediction, and segmentation.
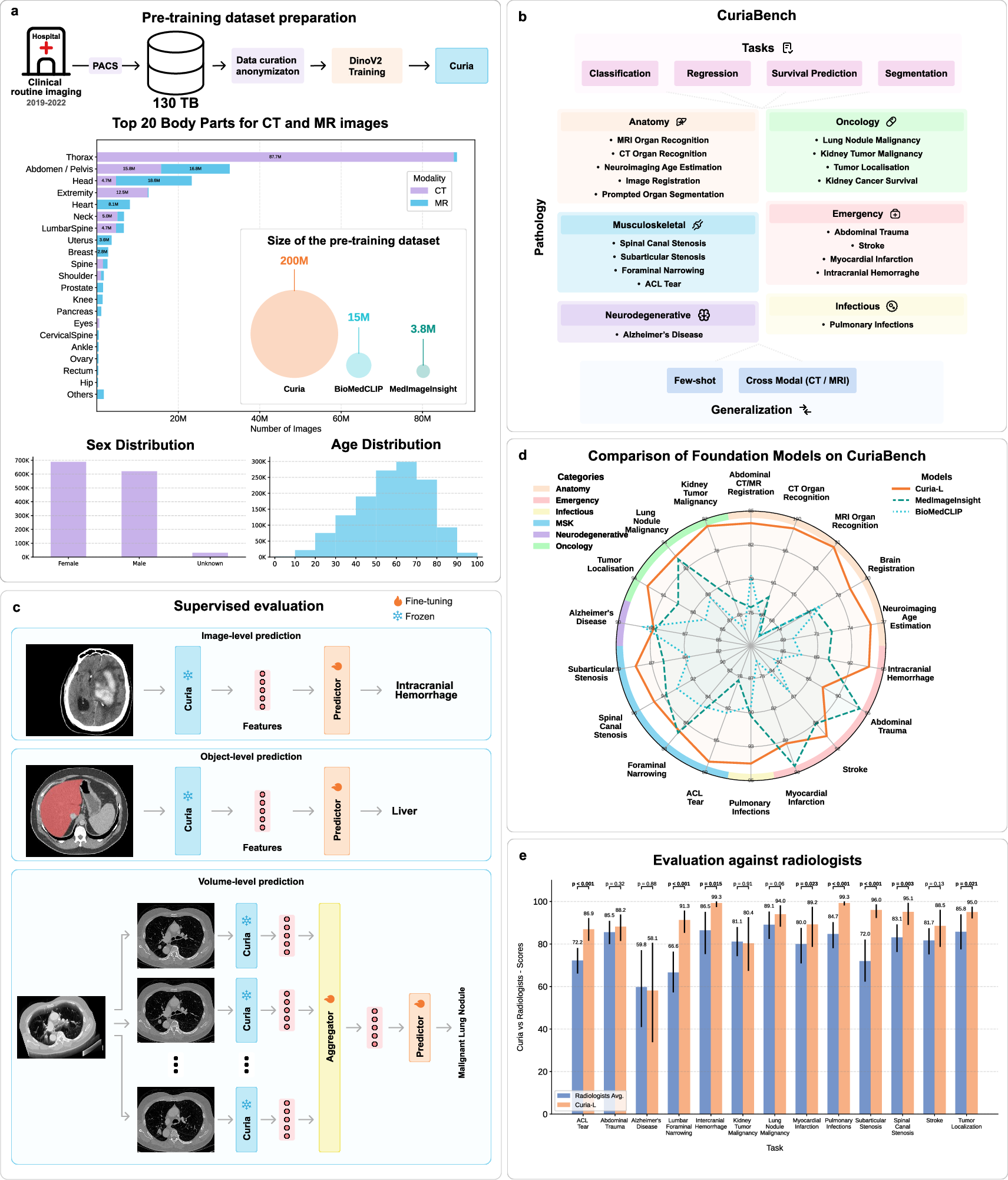
Figure 1: Overview of Curia, including pre-training methodology, benchmark tasks, evaluation strategies, and comparative performance against MedImageInsight, BioMedCLIP, and radiologists.
Model Architecture and Pre-Training
Curia employs the ViT-B and ViT-L architectures, corresponding to Curia-B (86M parameters) and Curia-L (300M parameters). Images are resized to 512×512 and tokenized into 16×16 patches. The DINOv2 self-supervised objective combines image-level and patch-level alignment between teacher and student networks, with regularization and cropping-based augmentations. Training is performed on distributed clusters of A100 GPUs, with Curia-L trained on the full 200M image dataset for 475,000 steps.
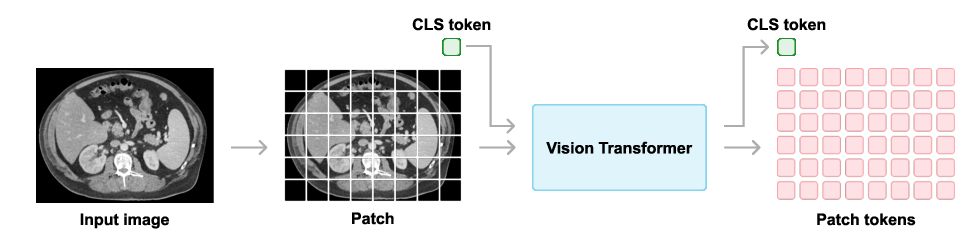
Figure 2: Vision Transformer architecture used in Curia, showing patch tokenization and class token for image-level tasks.
Benchmarking: CuriaBench
CuriaBench comprises 19 tasks across CT and MRI modalities, including anatomical classification, organ recognition, age estimation, image registration, prompted segmentation, oncology (tumor localization, malignancy, survival), musculoskeletal, emergency, neurodegenerative, and infectious disease detection. Each task is evaluated with appropriate metrics (accuracy, AUC, balanced accuracy, r2, c-index) and compared against MedImageInsight, BioMedCLIP, and radiologist performance.
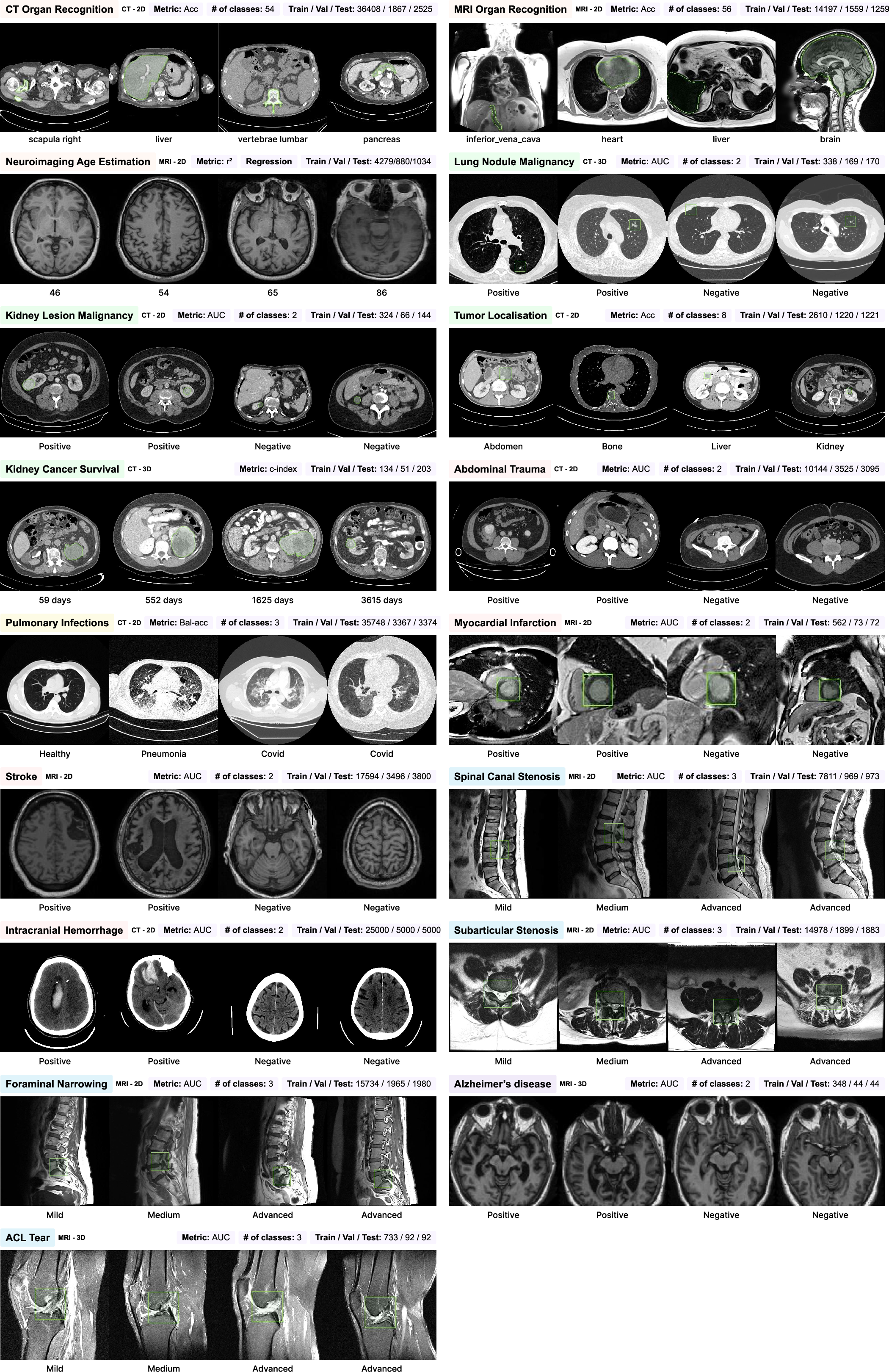
Figure 3: List of downstream tasks in CuriaBench, detailing modality, type, metric, class count, and dataset sizes.
Anatomical and Cross-Modality Generalization
Curia-L achieves near-perfect accuracy (98.4%) in CT organ classification, outperforming MedImageInsight (88.2%) and BiomedCLIP (85.0%). On MRI, Curia-L reaches 89.1% accuracy, with competitors at 63.2%. In few-shot settings, Curia maintains high accuracy with minimal labeled data, demonstrating strong data efficiency. Cross-modality experiments show Curia's balanced accuracy drop is only 9.2% (CT→MRI), while other models suffer drops of 35–72%. For MRI→CT, Curia's accuracy is virtually unchanged, indicating robust modality-agnostic anatomical representations.
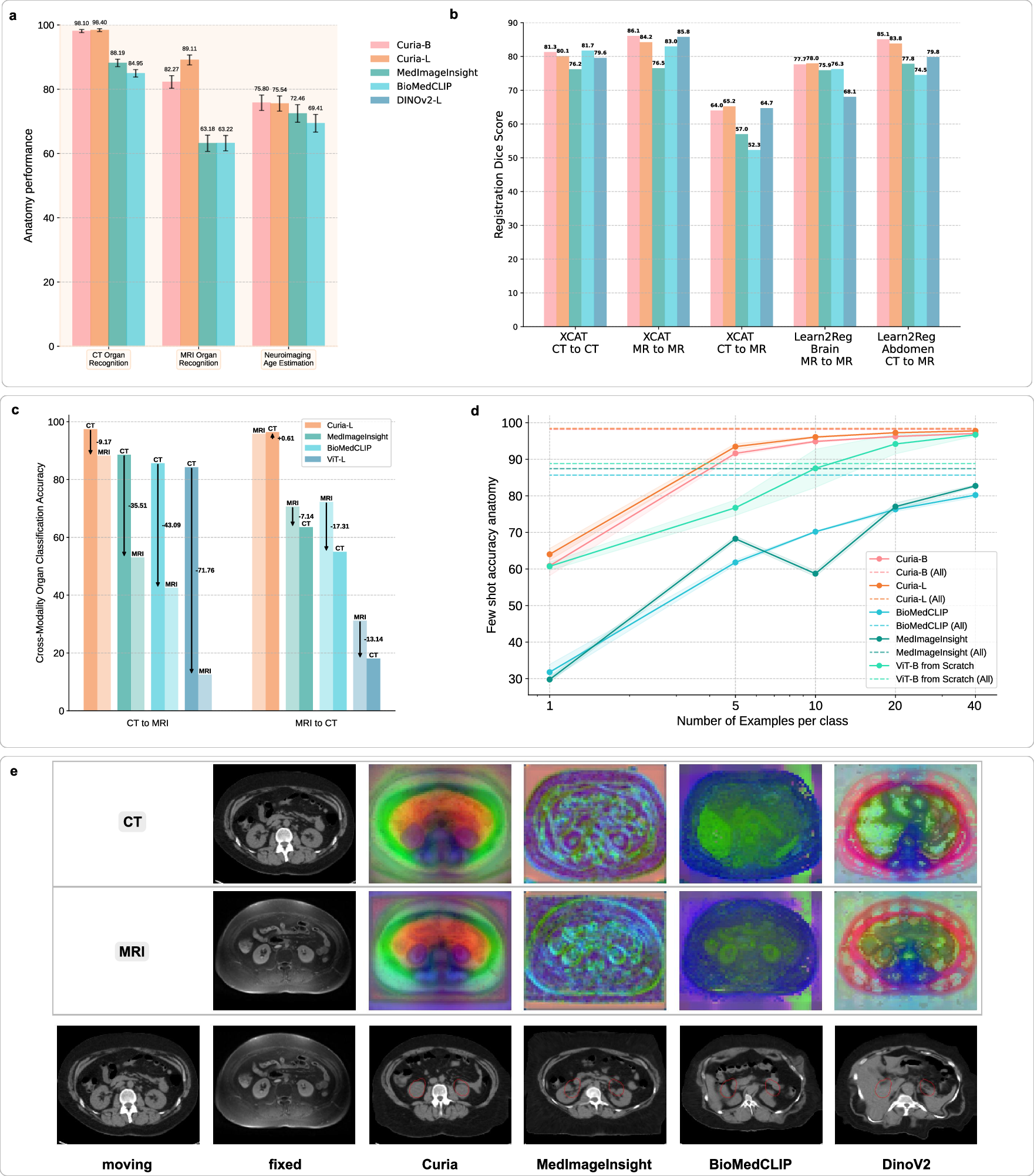
Figure 4: Comparative performance of foundation models on anatomical tasks, registration, cross-modality generalization, and data efficiency.
Image Registration and Feature Embedding
Curia outperforms or matches other FMs in zero-shot registration tasks (XCAT, Learn2Reg Abdomen, Learn2Reg Brain), with mean Dice scores up to 86.1% (MR-to-MR). Cross-modality registration (CT-to-MR) yields Dice scores of 65.3% (Curia-L), surpassing MedImageInsight and BiomedCLIP. PCA visualizations of feature maps reveal Curia's embeddings are more structurally diverse and discriminative, supporting its superior registration and generalization capabilities.
Prompted Segmentation
Curia is adapted for prompted segmentation by replacing the SAM encoder with Curia's backbone and fine-tuning the prompt encoder and mask decoder. On the AMOS dataset, Curia-L achieves Dice scores of 91.5% (bounding box) and 87.5% (point prompt), matching or exceeding RadSAM and outperforming SAM (70.2% and 33.0%, respectively). Qualitative results show Curia-L accurately segments disconnected liver components with minimal prompts.
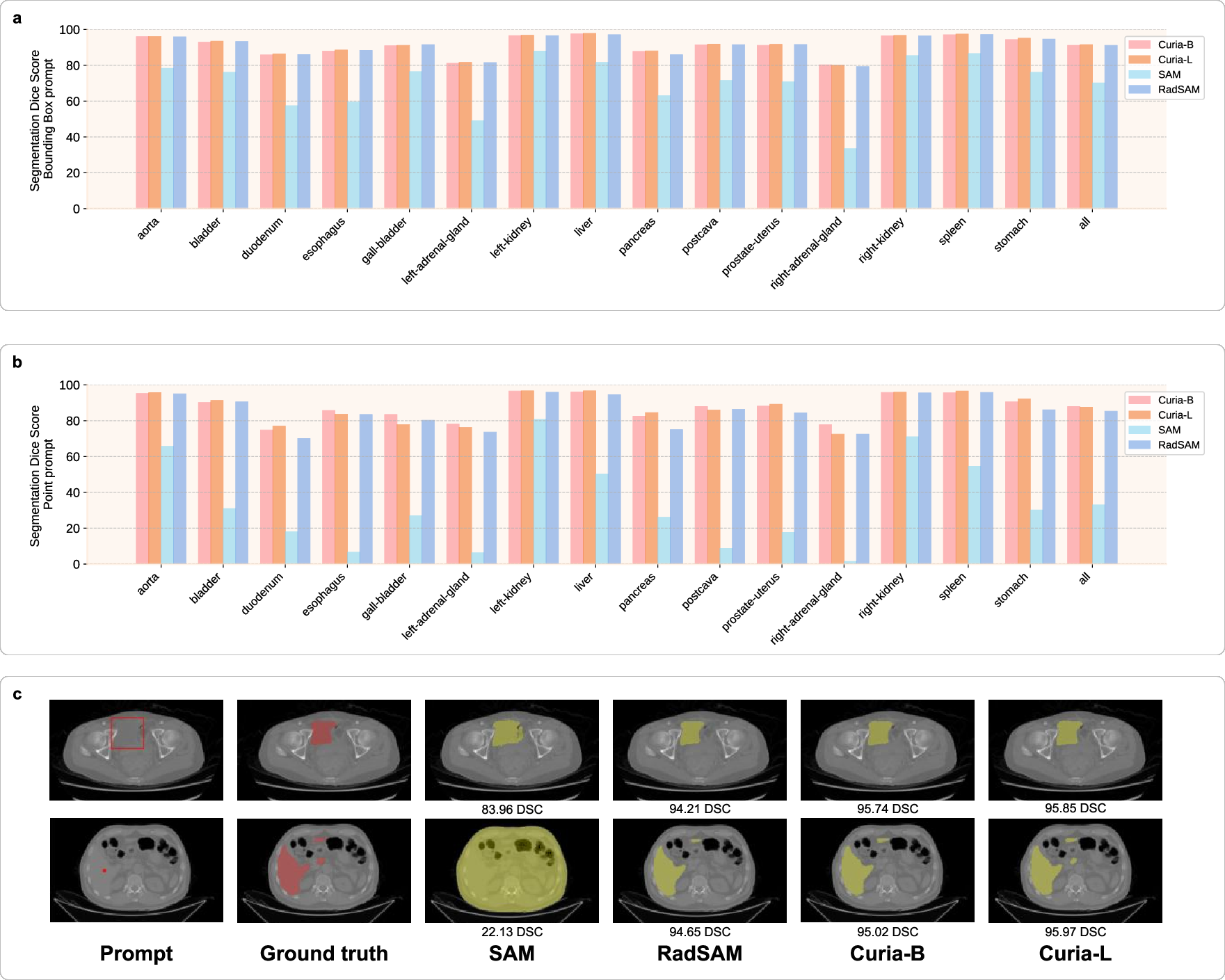
Figure 5: Prompted segmentation performance of Curia, SAM, and RadSAM, including quantitative Dice scores and qualitative segmentation maps.
Oncology Tasks
Curia-L attains balanced accuracy of 91.7% in tumor localization, outperforming MedImageInsight and BiomedCLIP. For kidney lesion malignancy, Curia-L achieves AUROC of 80.3%, surpassing BiomedCLIP (62.9%) and MedImageInsight (67.8%). In lung nodule malignancy, Curia-B reaches AUROC of 95.0%, exceeding Harvard Onco-FM's feature-based approach (88.2%) and matching its fine-tuned result (94.4%). Curia also demonstrates strong few-shot learning in oncology, with rapid performance gains as labeled data increases.
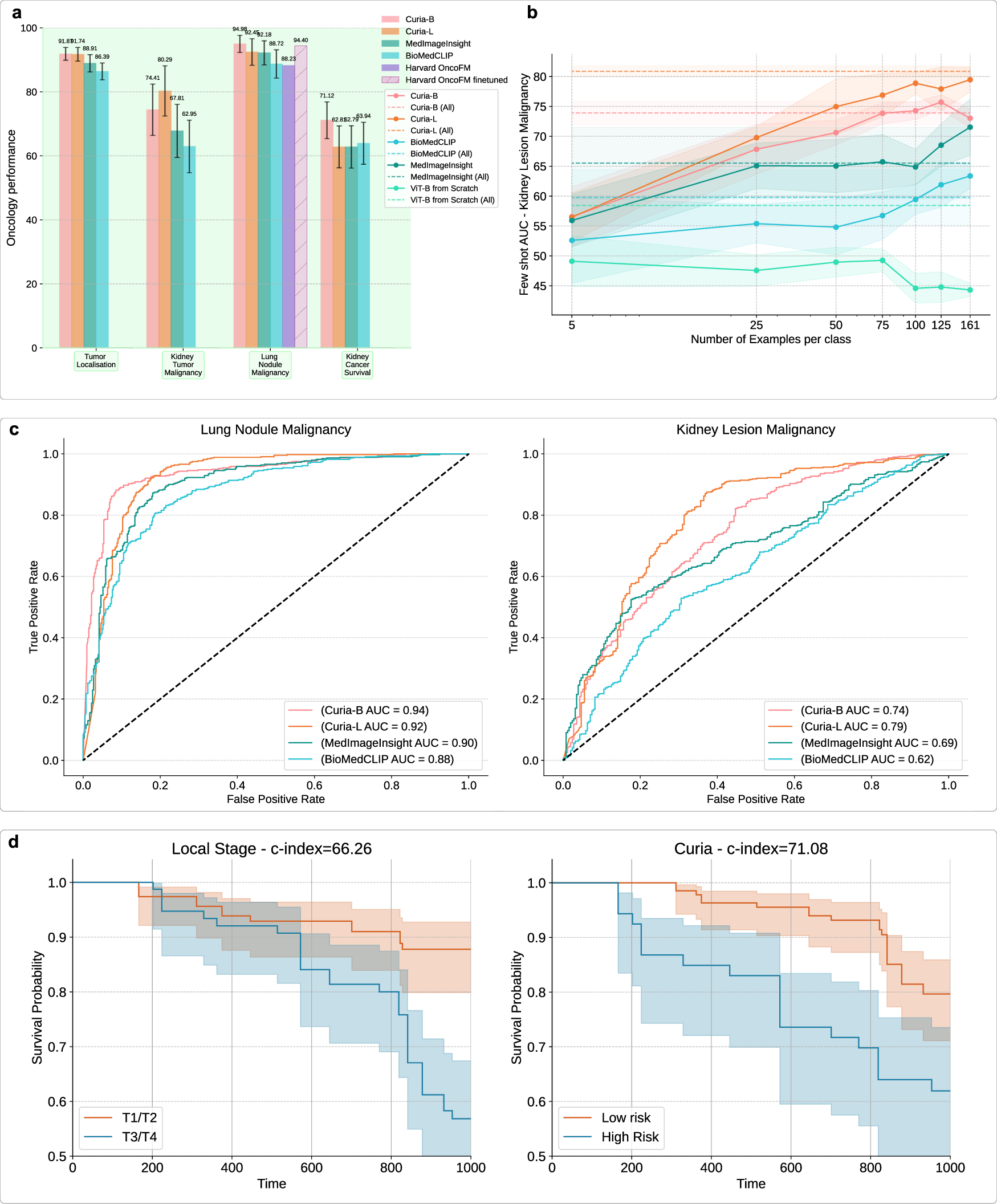
Figure 6: Curia's performance on oncology tasks, including tumor site classification, malignancy prediction, survival analysis, and ROC curves.
Survival Prediction
Curia-B achieves a c-index of 0.71 for renal malignancy survival prediction, outperforming BiomedCLIP (0.64), MedImageInsight (0.63), and the clinical T-stage (0.66). Kaplan-Meier curves stratified by Curia's risk predictions show clear separation, indicating clinically relevant prognostic capabilities.
Musculoskeletal, Emergency, Neurodegenerative, and Infectious Tasks
Curia-L delivers leading AUROC scores in musculoskeletal disease assessment (up to 93.7%), ACL tear detection (87.3%), intracranial hemorrhage (93.5%), and pulmonary infection detection (93.4%). In neurodegenerative disease (Alzheimer's), Curia-B and Curia-L achieve AUROC of 87.8% and 87.7%, respectively, matching or exceeding competitors.
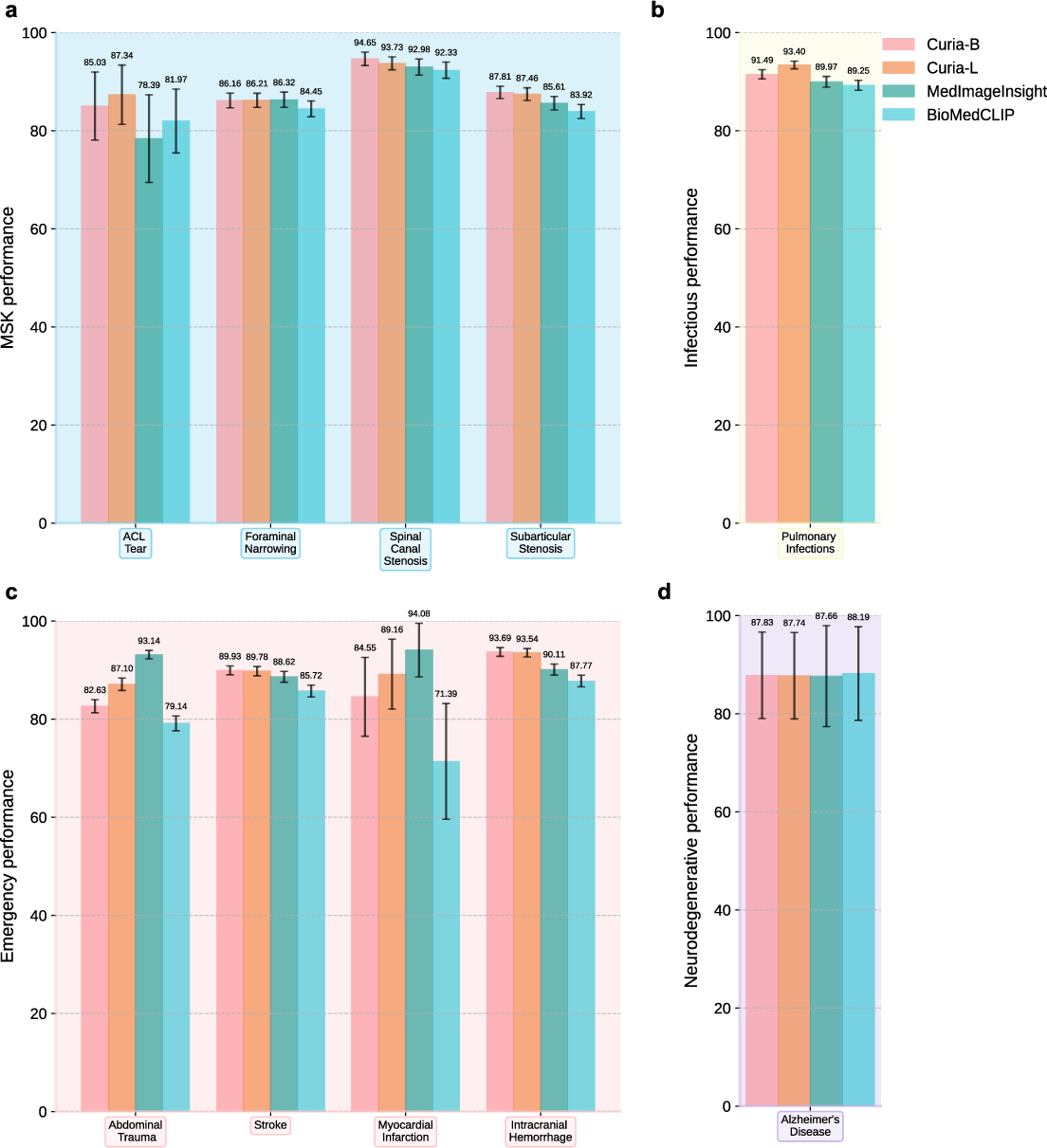
Figure 7: Performance comparison of Curia, BioMedCLIP, and MedImageInsight across musculoskeletal, infectious, emergency, and neurodegenerative subsets.
Comparison to Radiologists
Curia's performance is comparable to, and often exceeds, that of final-year radiology residents across 14 imaging tasks. This is supported by robust bootstrapped confidence intervals and paired hypothesis testing.
Interpretability and Feature Analysis
Attention map visualizations show Curia's decision-making is anatomically focused, with negative cases yielding diffuse attention and positive cases highlighting relevant regions. Keypoint matching experiments demonstrate Curia's ability to transfer features across modalities and patients, supporting its anatomical consistency and generalization.
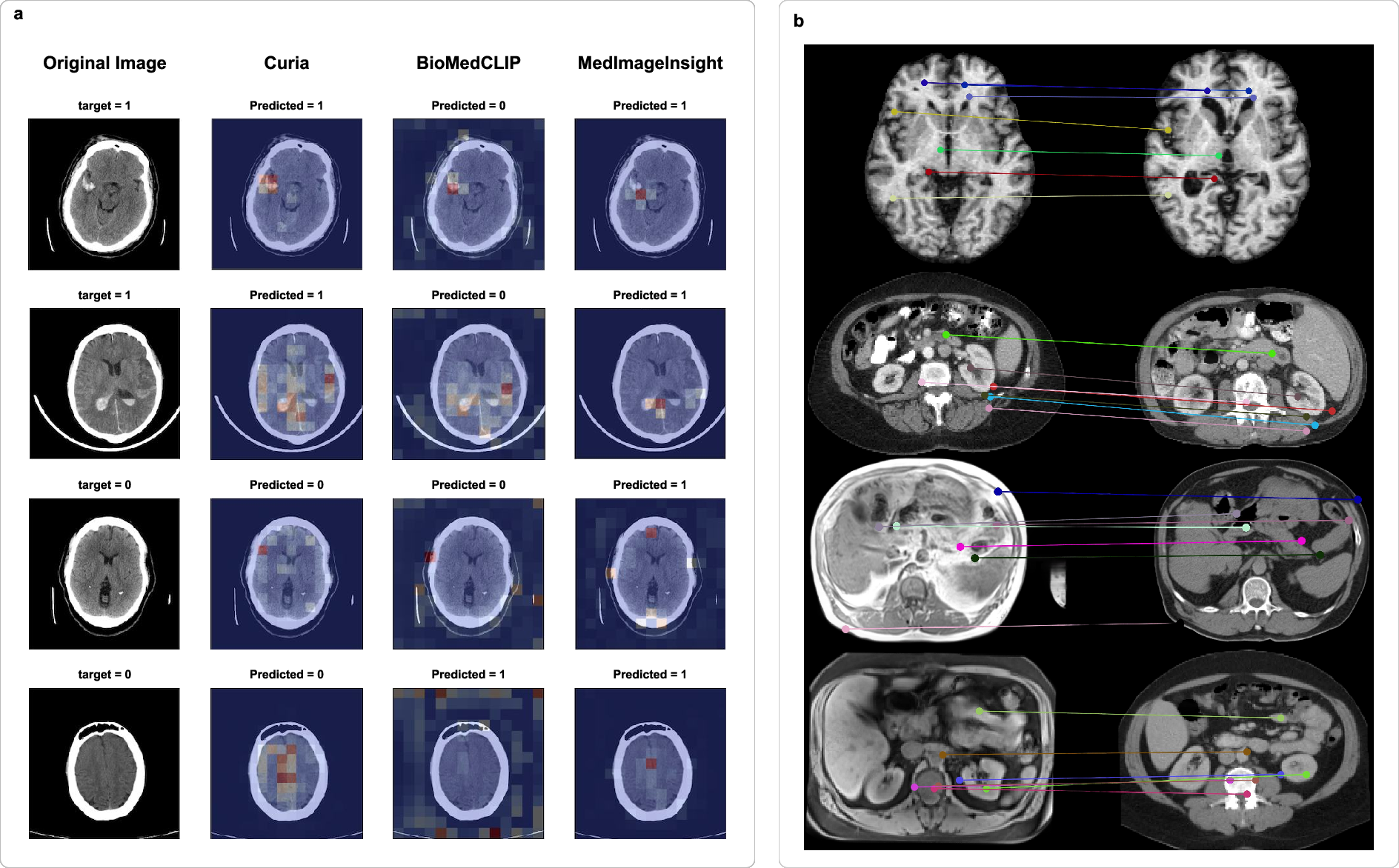
Figure 8: (a) Attention maps for intracranial hemorrhage classification; (b) Keypoint matching across modalities and patients.
Scaling Laws
Scaling experiments confirm that both dataset size and training duration are critical for downstream performance. ViT-B and ViT-L models show monotonic improvements with increased data and steps, with diminishing returns at extreme scales.
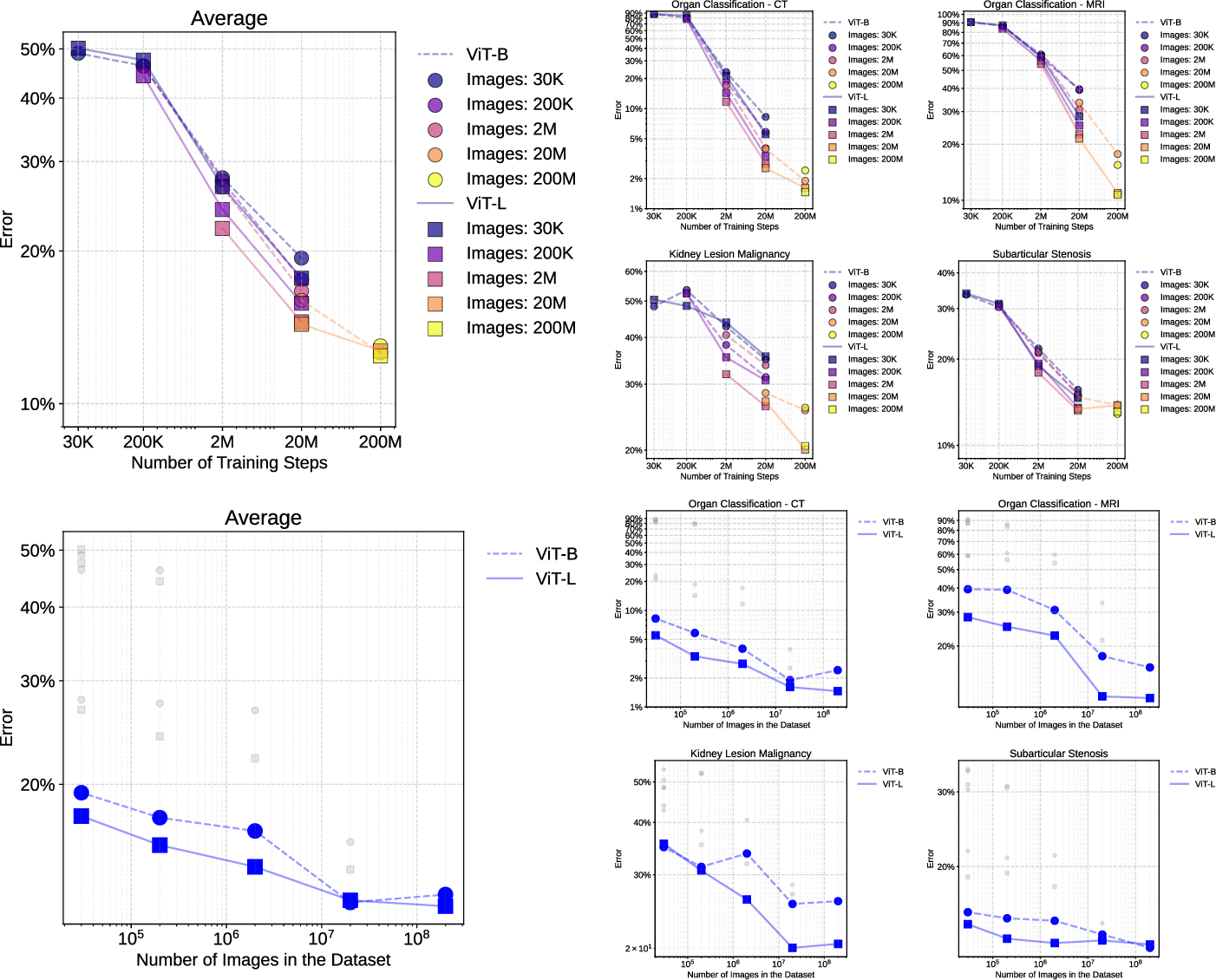
Figure 9: Scaling curves for ViT-B and ViT-L architectures, showing performance trends across dataset sizes and training steps.
Implementation Details
Curia is implemented in PyTorch, leveraging the DINOv2 codebase with modifications for medical imaging. DistributedDataParallel is used for multi-GPU training. Downstream tasks are adapted via lightweight heads (linear, MLP, attention-based pooling) trained on frozen backbone features. For segmentation, a two-stage fine-tuning protocol is employed. Zero-shot registration follows DINO-Reg methodology, utilizing class and patch tokens for rigid and deformable alignment. All experiments are reproducible on standard HPC clusters and consumer GPUs.
Limitations and Future Directions
Curia's training data is sourced from a single institution, potentially introducing site-specific biases. The model operates on 2D slices, requiring aggregation strategies for volumetric tasks; native 3D FMs may offer further improvements. The benchmark does not yet cover ultrasound, X-ray, or nuclear medicine. Clinical integration requires addressing regulatory, workflow, and IT system challenges.
Future work should focus on expanding modality coverage, developing native 3D architectures, and integrating multimodal data (e.g., EHR, textual reports) to enable richer contextual understanding and conversational interfaces.
Conclusion
Curia establishes a new standard for radiological foundation models, demonstrating robust generalization, data efficiency, and cross-modal capabilities across a diverse set of clinical tasks. Its performance matches or exceeds that of radiologists and prior FMs, with strong numerical results in anatomical, oncological, and emergency imaging. The release of Curia's weights and CuriaBench provides a foundation for further research and clinical translation. The evolution of Curia and similar models will likely involve deeper multimodal integration and enhanced interpretability, driving progress toward universal, data-efficient AI in medical imaging.










