- The paper introduces MedDINOv3, which adapts ViT-based foundation models using multi-scale token aggregation to significantly improve segmentation performance (e.g., DSC from 78.39% to 85.51% on AMOS22).
- It employs a three-stage, domain-adaptive pretraining pipeline on a diverse CT-3M dataset to bridge the gap between natural and medical imaging domains.
- Comparative evaluations show MedDINOv3 outperforms both CNN-based and transformer baselines across four public segmentation benchmarks, demonstrating state-of-the-art results.
MedDINOv3: Adapting Vision Foundation Models for Medical Image Segmentation
Introduction
MedDINOv3 presents a systematic approach for adapting large-scale self-supervised vision foundation models, specifically DINOv3, to the domain of medical image segmentation. The work addresses two persistent challenges: the underperformance of ViT-based backbones compared to specialized CNNs in dense prediction tasks, and the substantial domain gap between natural and medical images that impedes direct transfer of pretrained representations. The authors propose architectural refinements to plain ViTs and introduce a multi-stage, domain-adaptive pretraining pipeline leveraging a curated dataset of 3.87M CT slices (CT-3M). MedDINOv3 demonstrates state-of-the-art performance across four public segmentation benchmarks, providing evidence for the viability of vision foundation models as unified backbones in medical imaging.
Architectural Refinements for Medical Segmentation
The baseline architecture utilizes a DINOv3 ViT-B encoder paired with a lightweight Primus-style decoder, minimizing convolutional influence to maximize the impact of transformer representations. The authors identify that using only the final transformer block as input to the decoder limits the model's ability to capture hierarchical spatial priors, which are critical for dense segmentation. To address this, MedDINOv3 aggregates patch tokens from multiple intermediate layers (blocks 2, 5, 8, 11), concatenating them to enrich spatial context for the decoder. This multi-scale token aggregation yields a substantial improvement in segmentation accuracy.
High-resolution training is another key refinement. Rather than reducing patch size, which increases computational overhead, the model resamples axial slices to thinner spacing and maintains a high input resolution (896×896), preserving local anatomical details. These architectural modifications collectively raise the ViT-B performance on AMOS22 from 78.39% to 85.51% DSC.
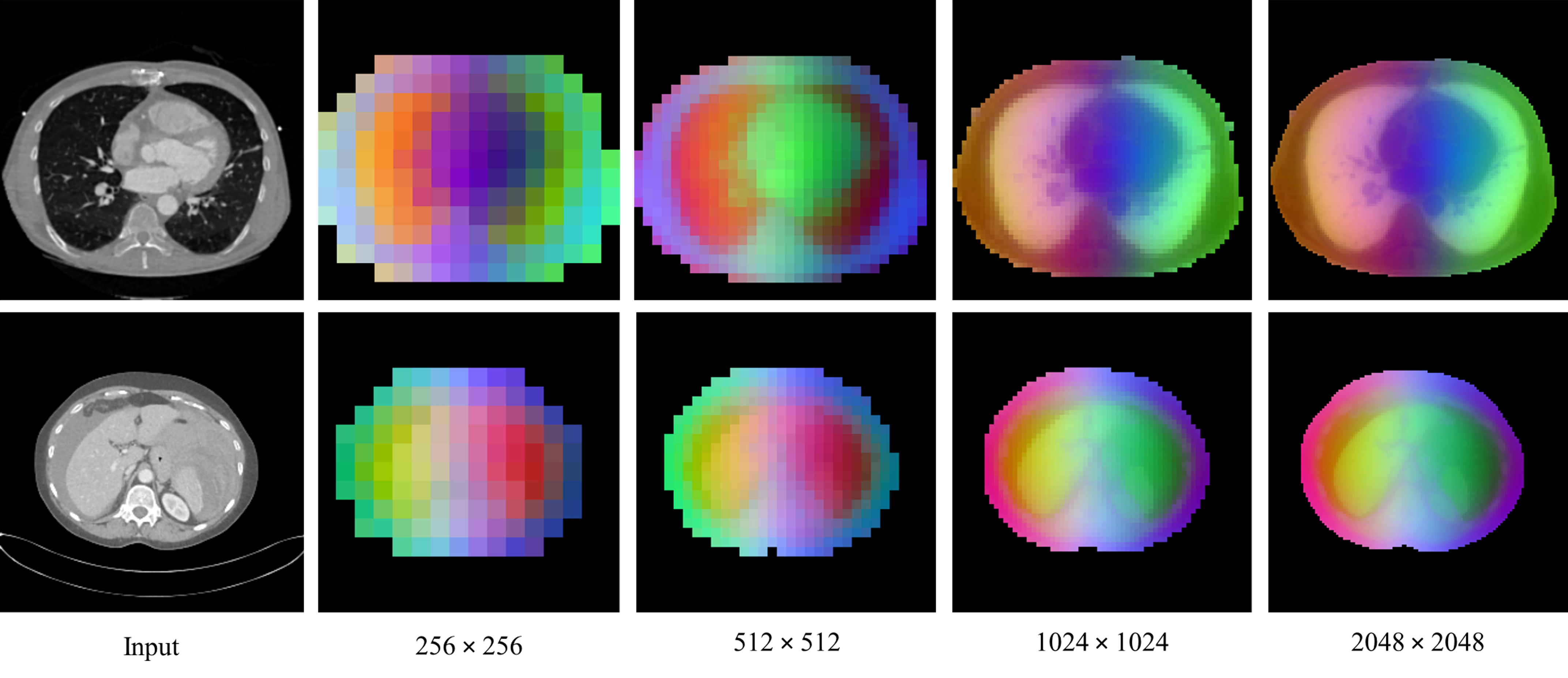
Figure 1: MedDINOv3 PCA maps at progressively higher resolution, visualizing dense features and focusing on CT foreground.
Domain-Adaptive Pretraining on CT-3M
To bridge the domain gap, MedDINOv3 is pretrained on CT-3M, a large-scale, heterogeneous collection of axial CT slices from 16 datasets, covering over 100 anatomical structures. The pretraining follows a three-stage DINOv3 recipe:
- Stage 1: DINOv2-style self-distillation and patch-level latent reconstruction, enforcing global-local crop invariance and learning local patch correspondence.
- Stage 2: Gram anchoring, regularizing the Gram matrix of patch features to prevent collapse and maintain patch-level consistency. The Gram teacher operates on higher-resolution crops, and its feature maps are downsampled to match the student.
- Stage 3: High-resolution adaptation, mixing global and local crops of various resolutions and retaining gram anchoring to stabilize patch similarity structures.
Ablation studies reveal that stage 1 pretraining provides the most significant boost in segmentation performance, while gram anchoring (stage 2) yields only marginal gains, likely due to minimal patch degradation observed during stage 1. High-resolution adaptation (stage 3) further improves dense feature quality and segmentation accuracy.
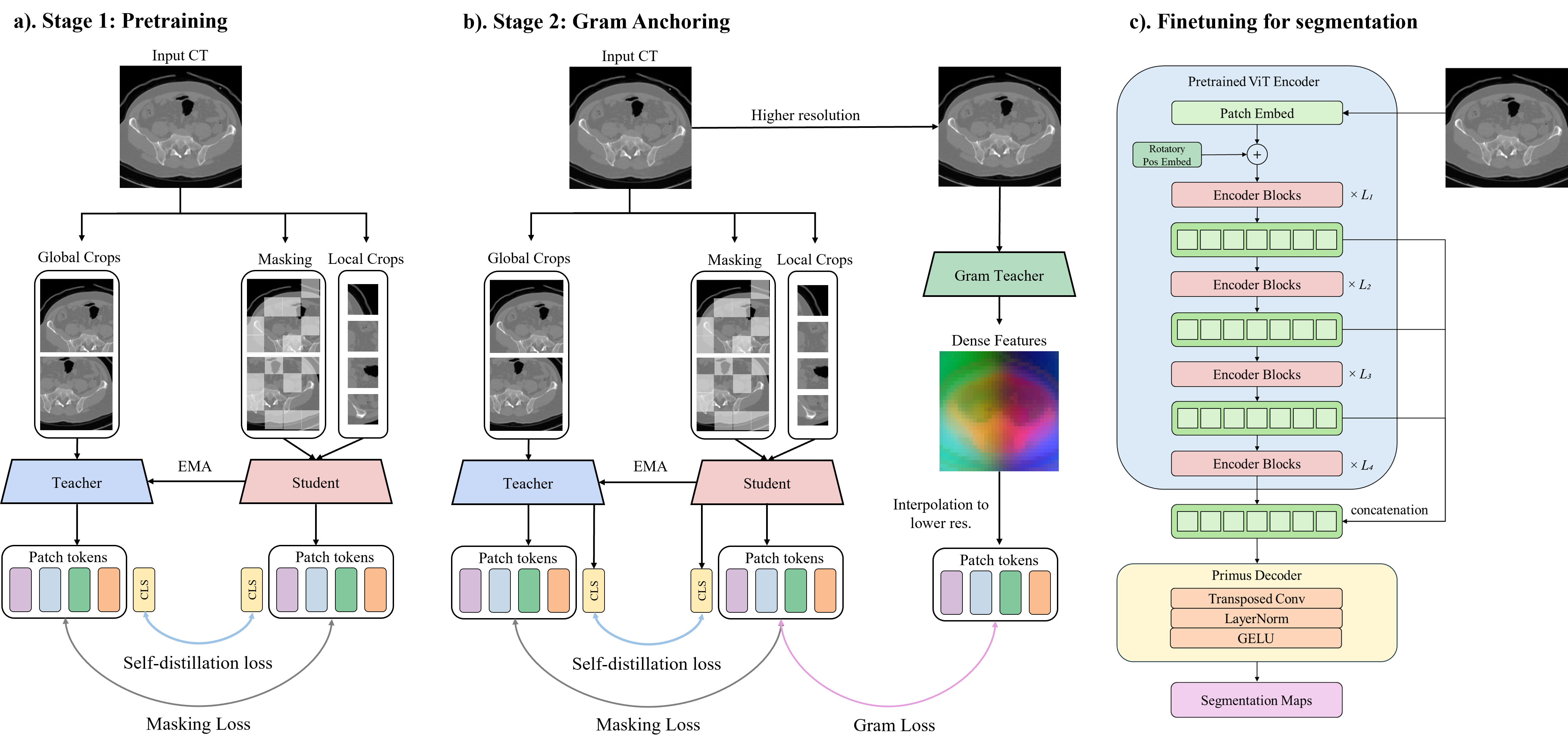
Figure 2: Overall framework of MedDINOv3, illustrating the three-stage pretraining pipeline and finetuning for segmentation.
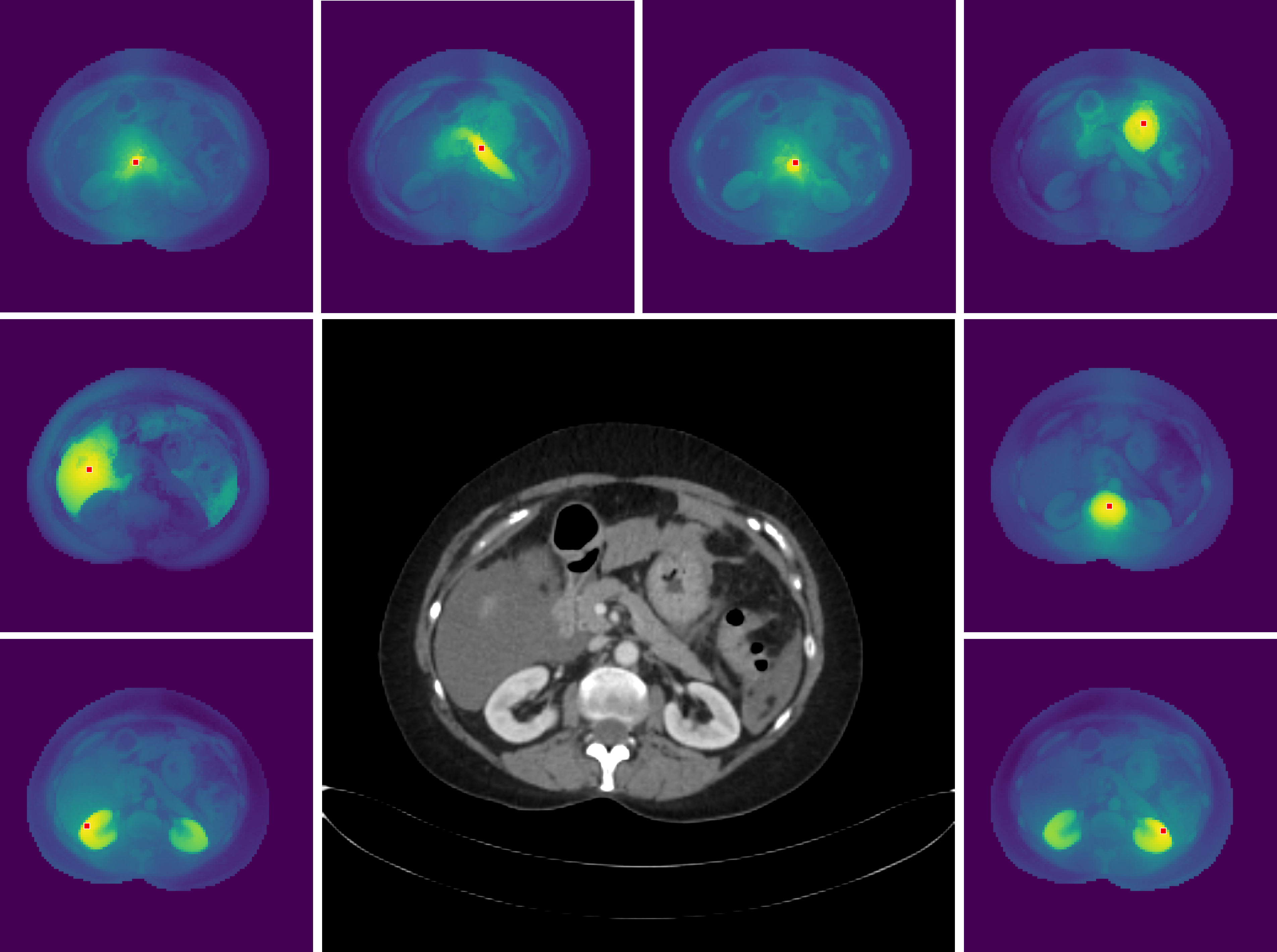
Figure 3: High-resolution dense features of MedDINOv3, visualized via cosine similarity maps between reference and all other patches.
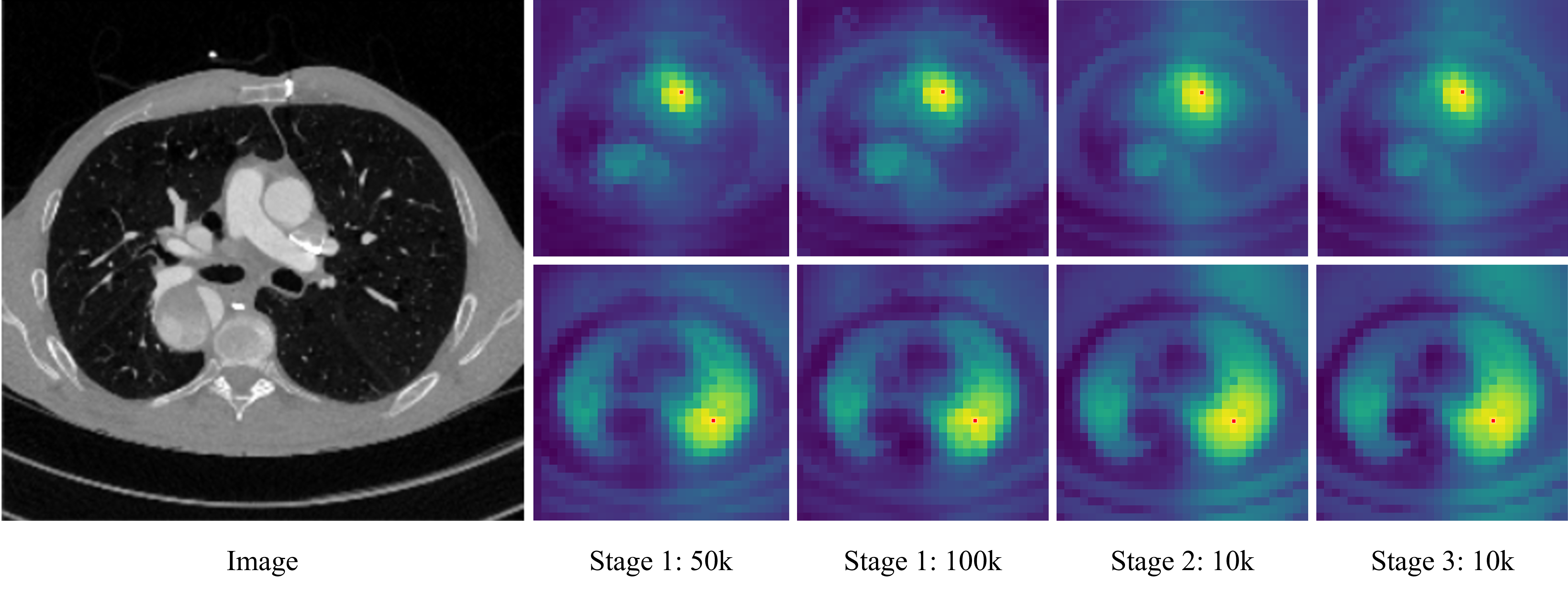
Figure 4: Evolution of cosine similarity between reference patch and all other patches, showing no severe patch degradation in stage 1.
Experimental Results and Comparative Analysis
MedDINOv3 is evaluated on four public benchmarks: AMOS22, BTCV, KiTS23, and LiTS, encompassing both organ-at-risk and tumor segmentation tasks across CT and MRI modalities. The model is compared against strong CNN (nnU-Net) and transformer (SegFormer, DINO U-Net) baselines. MedDINOv3 consistently outperforms nnU-Net in organ segmentation (+2.57% DSC on AMOS22, +5.49% DSC on BTCV) and achieves competitive results in tumor segmentation (70.68% DSC on KiTS23, 75.28% DSC on LiTS). SegFormer and DINO U-Net lag behind, underscoring the importance of domain-adaptive pretraining and architectural refinements.
The ablation paper confirms that DINOv2-style pretraining and high-resolution adaptation are the primary contributors to performance gains, while gram anchoring is optional in this setting. MedDINOv3 maintains strong boundary accuracy (NSD) for organ segmentation and matches CNN performance on tumor datasets.
Implications and Future Directions
MedDINOv3 demonstrates that simple ViT-based architectures, when paired with targeted domain-adaptive pretraining, can close the performance gap with, and in some cases surpass, specialized CNNs in medical image segmentation. The findings suggest that the weak locality bias of ViTs can be mitigated through multi-scale token aggregation and high-resolution training. The marginal utility of gram anchoring in this context indicates that patch-level consistency is largely preserved with DINOv2-style objectives on large-scale medical data.
Practically, MedDINOv3 offers a unified, scalable backbone for diverse medical segmentation tasks, reducing the need for highly specialized architectures. The approach is extensible to other modalities (e.g., MRI, ultrasound) and tasks (e.g., detection, registration) with appropriate domain-adaptive pretraining. Future work may explore further architectural simplification, integration with multimodal foundation models, and efficient adaptation strategies for low-resource settings.
Conclusion
MedDINOv3 provides a robust framework for adapting vision foundation models to medical image segmentation, combining architectural refinements and domain-adaptive pretraining. The model achieves state-of-the-art performance on multiple benchmarks, demonstrating that ViT-based backbones, when properly adapted, are competitive with or superior to established CNNs. The work highlights the importance of large-scale, heterogeneous medical datasets and systematic pretraining strategies in transferring foundation model capabilities to radiology. MedDINOv3 sets a precedent for future research in unified, generalizable medical vision models.



