- The paper presents a comprehensive review of foundation models in pathology, radiology, and ophthalmology, emphasizing the shift to self-supervised pretraining over traditional methods.
- The paper details advanced methodologies such as ViT-based architectures, contrastive and generative SSL objectives, and parameter-efficient adaptation strategies to improve clinical translation.
- The paper underscores challenges in data curation, model explainability, and bias while outlining a future roadmap for enhancing clinical deployment of foundation models.
Foundation Models in Medical Imaging: Review, Methodological Advances, and Prospects
Introduction
The paper "Foundation Models in Medical Imaging – A Review and Outlook" (2506.09095) provides a systematic and granular review of the development and deployment of foundation models (FMs) across pathology, radiology, and ophthalmology. The authors scrutinize over 150 works, construct an integrative framework to dissect the technical pillars underpinning successful FM pipelines, and contrast the downstream clinical applications and open challenges across the primary medical imaging domains. The principal focus is a rigorous technical synthesis of model architectures, self-supervised learning (SSL) objectives, and adaptation protocols that distinguish high-performing foundation models in the medical imaging ecosystem.
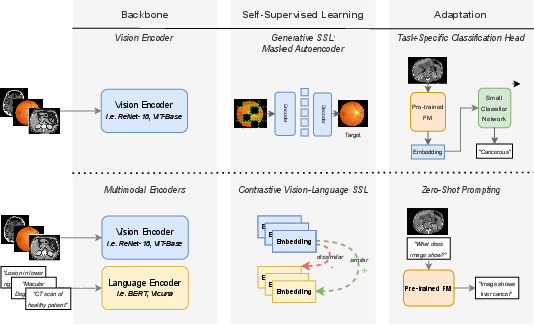
Figure 1: Schematic overview of two FM paradigms: single-modal masked autoencoding (top), and dual-modal vision-language contrastive training (bottom), each combined with tailored adaptation mechanisms for downstream tasks.
Technical Framework: FM Building Blocks and Their Instantiation
Large-Scale Architectures and Pretraining Strategies
The FM paradigm in medical imaging is defined by large-scale encoders trained on massive, predominantly unlabeled datasets. The paper delineates the technical trade-offs between convolutional neural networks (CNNs) and transformers (notably Vision Transformers, ViT), clearly identifying the scaling limitations of CNNs due to inductive bias and describing how ViTs—despite lacking architectural priors—excel when massive data and compute are available. There is a strong trend toward ViT-based backbones or hybrid CNN-ViT constructs to harness spatial hierarchies and global attention. The largest published pretraining efforts (e.g., Virchow, UNI, Atlas, Phikon-v2) operate on up to 3.1M WSIs or 1.9B image patches in pathology.
Self-Supervised Learning Objectives
The authors provide a thorough account of SSL objectives, emphasizing that contrastive learning (SimCLR, MoCo, SwAV, BYOL, DINO/DINOv2, iBOT) and generative masked modeling (MAE, BEiT, I-JEPA) are the primary driving forces in FM pretraining. The distinction is made between contrastive approaches' reliance on informative negative pairs or teacher soft labels, and generative methods' semantic representation learning via reconstruction or embedding prediction. Vision-language FMs (VLFMs) extend this with cross-modal contrastive losses (as in CLIP, ALIGN, CoCa, etc.), and emerging multimodal pretraining protocols (e.g., late/early/mid fusion) optimize for cross-domain generalization.
Adaptation Protocols and Downstream Task Transfer
The adaptation spectrum spans non-parametric (e.g., linear probing, k-NN, prompt engineering for zero/few-shot inference) to parameter-efficient fine-tuning (LoRA, adapters, prompt tuning) and full model updates. The specific adaptation choice is modulated by the task's complexity, data availability, and compute constraints. Notably, parameter-efficient adaptation is crucial to democratize FM usage across resource-constrained academic and clinical environments.
Pathology Foundation Models: Data Scaling, SSL Evolution, and Multimodal Trends
Progression From Transfer Learning to In-Domain Self-Supervision
Early digital pathology relied on ImageNet transfer learning; contemporary FMs now ubiquitously leverage in-domain SSL, often on multi-organ, multi-stain, and multi-institutional training data. There is a bifurcation between tile-level FMs (e.g., SimCLR, REMEDIS, iBOT, DINOv2-based Phikon/v2, Virchow, UNI, Atlas) and slide-level FMs (LongViT, Prov-GigaPath, PRISM, THREADS), with the latter incorporating hierarchical context modeling or vision-language pretraining to capture spatial and biological context at scale.
SSL Methodological Advances & Scaling Effects
Contrastive methods initially dominated pathology FM training but show diminishing returns as semantic redundancy is not handled optimally. Recent evidence suggests that DINOv2 and iBOT-style self-distillation and masked embedding objectives outperform older contrastive schemes when trained at scale. Large model and data scale correlates with improved downstream benchmarks, although the effect plateaus when data curation or augmentational diversity is insufficient—a finding directly contradicting naive scaling laws. E.g., Atlas (632M params; 520M patches), using high tissue, stain, scanner, and magnification diversity, equals or exceeds larger models trained on massive proprietary datasets.
Multimodal and Vision-Language Pathology FMs
Rapid expansion is seen in VLFMs (PLIP, CONCH, PLUTO, PA-LLaVA, PathChat, QuiltNet-LLaVA, PRISM) utilizing cross-modal contrastive training or generative instruction-tuning for report generation, retrieval, and zero-shot subtyping/diagnosis. These models align tile- or slide-level embeddings with expert-written captions, enabling zero- and few-shot pathology VQA.
Radiology Foundation Models: Modality Gaps, 3D Scaling, and Vision-Language Fusion
X-Ray, CT/MRI, and Generalist FM Trends
X-ray (particularly chest X-ray) serves as the prime testbed due to public datasets and lower 3D complexity, evidenced by models like CheXZero, CXR-CLIP, MedCLIP, MaCo, and the instruction-tuned CheXAgent. For volumetric modalities, resource and annotation constraints limit the breadth of 3D FM pretraining. Nevertheless, models such as RadFM, M3D-LaMed, VISION-MAE, Rad-CLIP, and CT-CLIP leverage 3D ViT backbones and are starting to match or exceed supervised benchmarks for abnormality detection and segmentation, often using mixed 2D/3D or text-image pair pretraining.
Vision-Only Versus Vision-Language Pipelines
There is a split between VFM and VLFM efforts: vision-only models (RAD-DINO, RayDINO, VISION-MAE, DeSD, SMIT, Medical Transformer) demonstrate superior performance in spatially intensive tasks like segmentation and detection, while VL encoders excel in report generation and clinical Q&A. As in pathology, scaling up training data (notably for 3D data) is hindered by lack of comprehensive open datasets, complicating progress beyond 2D/2.5D slices. Furthermore, the large-scale deployment of transform-based 3D models is currently constrained by computational costs.
Ophthalmology Foundation Models: Data Diversity, Multimodality, and Vision-Only Emphasis
Specialized FMs and Data-Efficient Scaling
The field is distinguished by robust open datasets from non-invasive modalities (color fundus photography, OCT), enabling vision-only FMs such as RETFound, its data- and compute-efficient RETFound-Green, and multimodal EyeFound. RETFound has been shown to produce features predictive not only for classical retinal diseases but also for systemic conditions (e.g., heart failure, Parkinson's, myocardial infarction), thus demonstrating high cross-disease transfer and substantiating the value of large-scale SSL models over traditional supervised approaches.
Integration of Text, Synthetic Data, and Cross-Modal Expansion
FLAIR and DERETFound show that encoding expert knowledge via textual tags or captions further boosts FM performance, particularly in low-data regime or for rare conditions. VisionFM and EyeFound attempt to extend FMs across the diverse imaging spectrum (e.g., angiography, slit-lamp, ultrasound), with initial evidence of strong cross-modality transfer; however, most reported advances are still centered on fundus/OCT data.
Segment Anything Model (SAM) and Cross-Domain Adaptation
The Segment Anything Model, when directly applied to medical data, exhibits heterogeneous zero-shot performance: strong for large, well-circumscribed objects and underwhelming for convoluted instances. Pathology-centric efforts using SAM (direct, LoRA/adapter tuning, prompt-based fine-tuning) and extensions such as SAM-U, DeSAM, and All-in-SAM report moderate performance improvements and demonstrate that domain adaptation or targeted prompt mechanisms are critical for cross-domain generalizability.
Technical and Clinical Challenges: Plateau Effects, Scalability, and Fairness
Data Curation, Open Clinical Datasets, and Evaluation Protocols
Scaling alone is not universally beneficial—performance gains plateau when semantic or biological diversity is exhausted. For pathology, curated datasets with heterogeneous tissue, stain, and scanner characteristics outperform brute-force scaling (as shown by RudolfV and Atlas). The lack of large, representative, open clinical datasets—especially for 3D radiology and rare ophthalmic disorders—remains a persistent barrier for reproducibility and benchmarking.
Robustness, Explainability, and Bias
Evidence is mounting that robustness to domain shifts (inter-institutional data, different scanner/staining) is not simply a function of model size. Targeted distillation, domain-specific augmentations, and explicit confounder-correcting objectives are required. Fairness and bias—particularly against minority subpopulations or rare disease categories—continue to confound FM deployment, requiring dedicated evaluation (robustness/vulnerability indices) and mitigation (e.g., federated or privacy-preserving training, balanced fine-tuning cohorts).
Adaptation, Efficient Finetuning, and Clinical Integration
Parameter-efficient methods (e.g., LoRA, adapters, prompt tuning) are essential for resource-constrained clinical settings and to facilitate multitask deployment without full retraining. Additionally, the ongoing regulatory and real-world deployment challenges (e.g., explaining model predictions, reducing hallucination, ensuring continual relearning) are underlined as critical open problems.
Future Prospects and Theoretical Implications
The reviewed evidence demonstrates that in-domain self-supervised learning is fundamental for performant FM pipelines in medical imaging. However, future progress depends upon (1) constructing deeply curated, distributed, multi-cohort clinical datasets for robust pretraining, (2) innovative cross-modal SSL objectives (e.g., incorporating text, omics, graph contexts), (3) parameter-efficient, modular adaptation schemes suited to hierarchical and multi-task settings, and (4) rigorous, transparent quantitative evaluation frameworks for fairness, robustness, and clinical translatability.
The paper implicitly challenges the notion that foundation models are a panacea for all medical imaging tasks, instead advocating for tailored, domain-aware architectures, SSL objectives, and adaptation protocols. This is a critical insight likely to guide future research toward maximizing the clinical and technical utility of FMs in medicine while navigating the inherent complexities and ethical demands of medical data.
Conclusion
"Foundation Models in Medical Imaging – A Review and Outlook" systematically articulates the technical underpinnings, empirical trajectories, and challenges faced by FMs in digital pathology, radiology, and ophthalmology. The synthesis of architectural scaling, SSL regime selection, and adaptation methods—combined with an incisive critique of current scaling limits, robustness, and clinical transfer—serves as a foundational reference for researchers and practitioners driving technical innovation in medical image analysis. The outlook emphasizes that future advances will hinge on harmonizing scale with diversity, robust domain adaptation, and clinically relevant evaluation, ultimately informing the next generation of generalist and specialist FMs in healthcare.


