- The paper introduces a computational framework to measure aging dynamics using transformer-based modeling on single-cell RNA-seq data.
- It applies ergodic theory and Hopf decomposition to distinguish conservative and dissipative gene components, quantifying differential aging.
- Results reveal tissue-specific rates of gene entropy and aging, offering insights for personalized therapeutic interventions.
The Dissipation Theory of Aging: A Quantitative Analysis Using a Cellular Aging Map
Introduction
The study proposes a novel approach to understanding aging through the lens of dynamical systems, framing it as a dissipative process. Utilizing ergodic theory, the research decomposes cellular aging into conservative and dissipative components, suggesting aging as predominantly driven by dissipative forces that lead to deviation from homeostasis and increased entropy over time. To quantify these dynamics, a transformer-based model was applied to single-cell RNA-seq data, generating a cellular aging map that provides insights into the differential aging rates across tissues and cell types. The research contributes a new computational framework enabling the measurement of age-related changes with molecular resolution.
Theoretical and Computational Framework
Aging is conceptualized as a dynamical system, characterized by conservative forces that maintain homeostasis and dissipative forces that instigate deviation from recurrent states. The study leverages ergodic theory and the Hopf decomposition theorem to underscore the significant role of dissipation in aging. The transformer-based model, integrating gene expression with age as a token, is key to quantifying these dissipation dynamics and constructing the cellular aging map (Figure 1).

Figure 1: Overview of theoretical and computational framework (a) decomposition of aging manifold in time using Hopf decomposition into conservative and dissipative components (b) aggregation of 1528 single-cell RNA-seq datasets encompassing various age groups, tissues, and cell types (c) modeling of gene expression using a transformer-based model with metadata information and age token, (d) cellular aging map constructed from the output embedding of the model.
Cellular Aging Map Analysis
Age Prediction and Tissue-Specific Dynamics
The predictive accuracy of the transformer-based model in estimating chronological age reveals significant tissue-specific dynamics. For instance, analyses exhibit that certain tissues like the respiratory airway show high prediction accuracy, suggesting uniformity in aging dynamics. Conversely, tissues such as breast tissue display variable deviation from chronological age, indicating accelerated or decelerated aging (Figure 2).
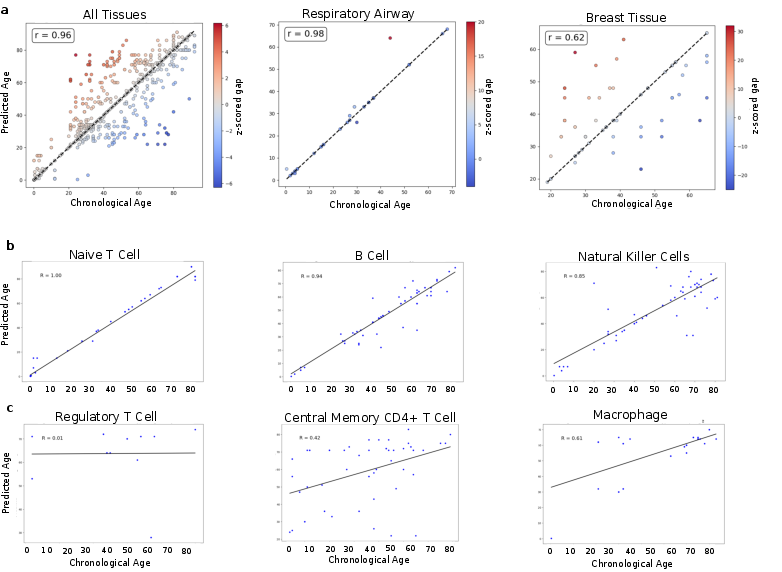
Figure 2: Construction of aging clock using model age prediction; deviations indicate cellular aging dynamics varying by tissue type.
Gene Dynamics and Dissipative Behaviors
Detailed embedding-based analyses uncover temporal and tissue-specific gene dynamics, highlighting the complex interplay between genes and aging across different life stages. Specific genes show temporal activity that correlates with aging in tissues like blood and lungs, evidencing their stage-specific roles in genetic regulation during aging (Figure 3).

Figure 3: Similarity analysis delineates tissue susceptibility to aging, identifying those heavily influenced by age and displaying dynamic gene activity.
Dissipation in Aging
Conservative vs. Dissipative Genes
Analysis using the Hopf decomposition distinguishes between conservative genes, maintaining stable embeddings, and dissipative genes showing high temporal drift. This classification aids in understanding aging's molecular mechanisms, driving differential aging trajectories. The research highlights how disease can amplify gene drift in dissipative genes, altering their steadiness observed in healthy conditions (Figure 4).
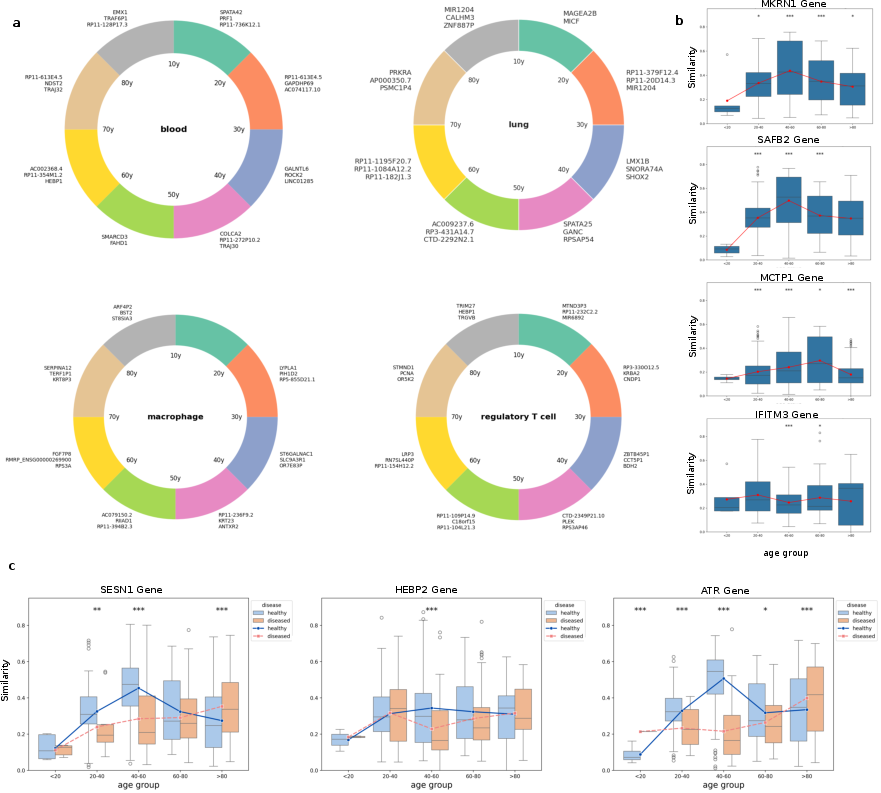
Figure 4: Embedding-based analysis highlights gene dynamics, showing stage-specific gene relevance and how disease alters gene trajectories.
Entropy as a Metric of Aging
Entropy analysis emerges as a robust metric for quantifying the complexity and variability in gene expression, aligning with the dissipative nature of aging. This study demonstrates that higher entropy is associated with greater uncertainty and molecular disorder in aging tissues, with disease exacerbating these effects (Figure 5).

Figure 5: Metrics of dissipation illustrate tissue-specific changes in entropy, highlighting differences in molecular organization between healthy and diseased states.
Discussion
The research introduces a comprehensive framework integrating theoretical underpinnings from dynamical systems with advanced modeling techniques to elucidate aging processes. By viewing aging through the lens of ergodic theory and the Hopf decomposition theorem, this study conveys aging as a dissipative process influenced by broader systemic dynamics rather than isolated molecular events. The insights garnered from this model challenge traditional chronological age metrics, calling for a focus on biological complexity and entropy as key indicators of aging. Future research should aim to integrate multi-omics data to further dissect the manifold complexity and refine the cellular aging map for broader applications in therapeutic strategies and personalized medicine.
Conclusion
The dissipation theory of aging provides a potent lens through which to view the complex dynamics of aging, offering a metric and model for understanding the process at a molecular level. This approach, leveraging the novel use of transformer-based models, opens avenues for new investigations into aging, potentially guiding therapeutic interventions aimed at mitigating age-related decline. This study is a step towards a robust interdisciplinary framework for aging research, combining the rigor of computational modeling with biological insights into the aging process.






