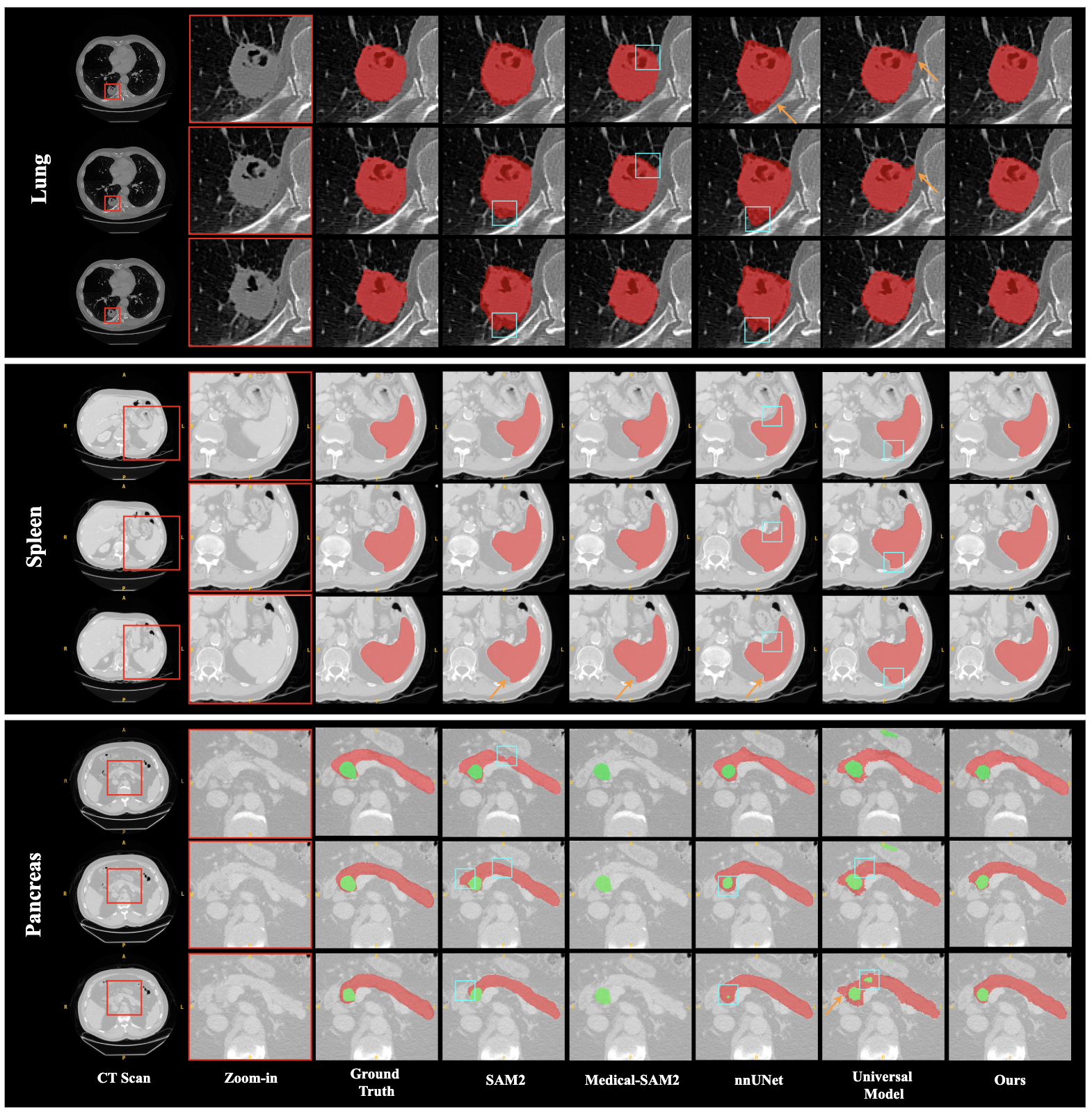
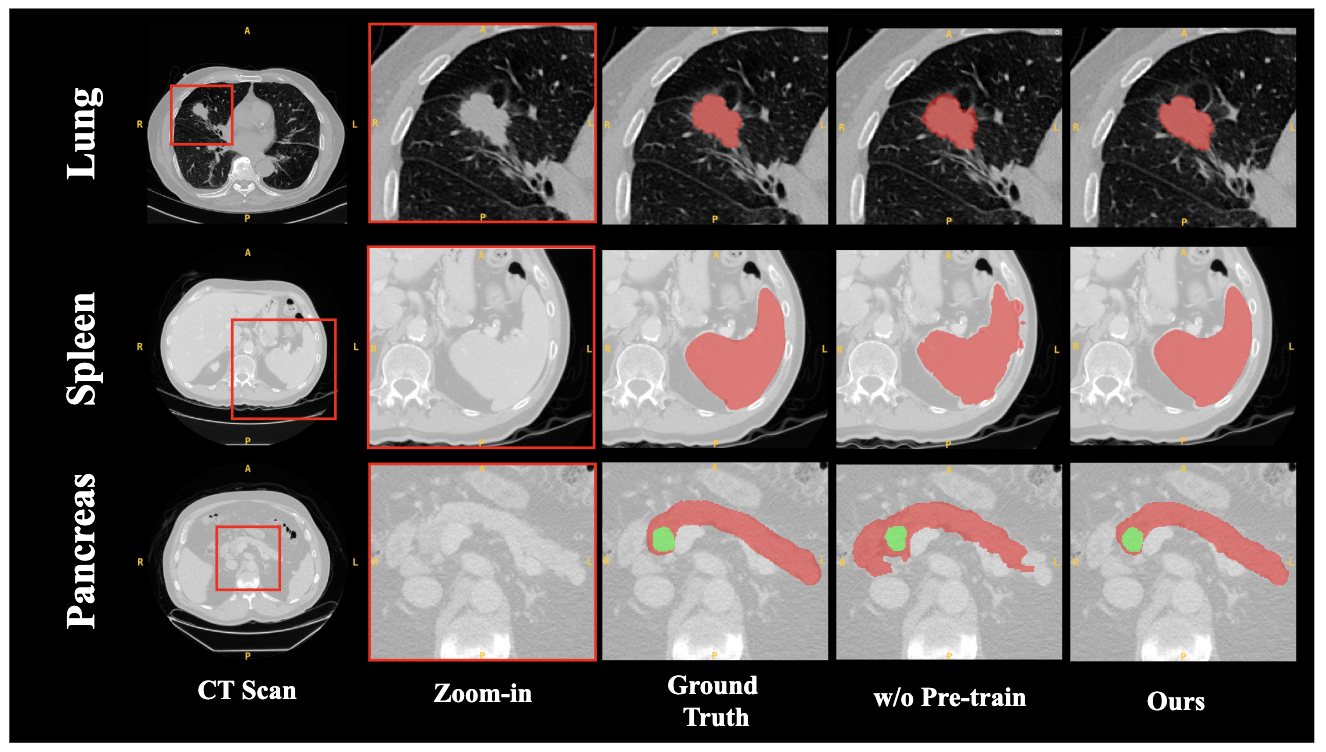
- The paper introduces SAM2-3dMed that leverages SRPP and BD modules to adapt SAM2 for enhanced 3D medical image segmentation.
- The paper employs a composite loss function combining Dice, MSE, and weighted binary cross-entropy to improve boundary delineation and volumetric accuracy.
- The paper demonstrates superior performance on MSD benchmark datasets, notably achieving significant Dice improvements on the Pancreas task.
SAM2-3dMed: Empowering SAM2 for 3D Medical Image Segmentation
Introduction
"Accurate segmentation of 3D medical images is imperative for applications such as disease diagnosis and treatment planning. The Segment Anything Model 2 (SAM2), renowned for its success in video object segmentation, faces challenges when applied to 3D medical images due to two domain discrepancies: bidirectional anatomical continuity and precise boundary delineation." To bridge these gaps, the paper introduces SAM2-3dMed as an adaptation of SAM2 specifically tailored for 3D medical imaging. The core contributions include a Slice Relative Position Prediction (SRPP) module and a Boundary Detection (BD) module, which respectively model inter-slice dependencies and enhance boundary precision.
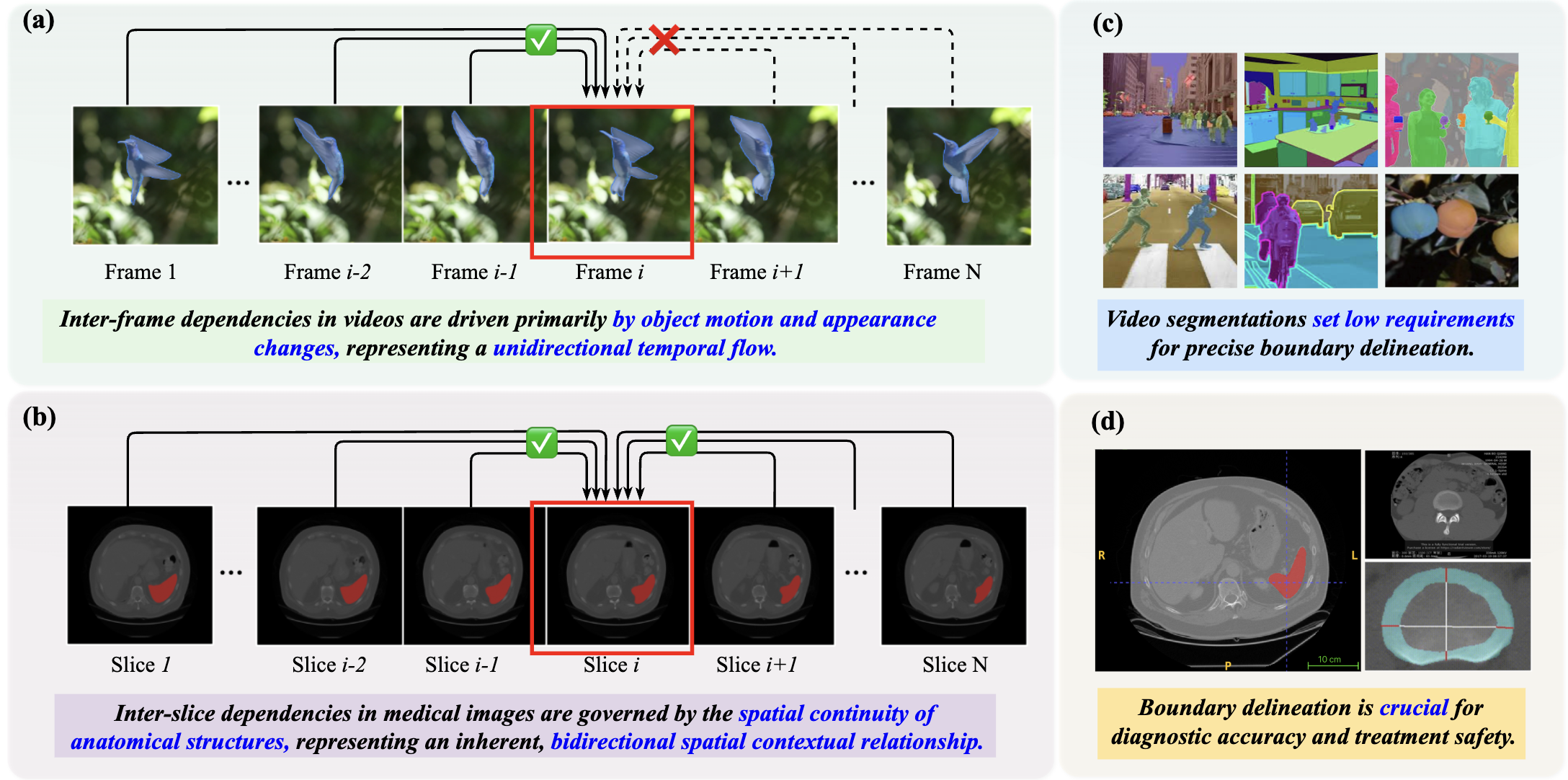
Figure 1: The comparison focuses on inter-frame dependencies in videos (a) vs. inter-slice dependencies in 3D medical images (b), and the importance of boundary segmentation for videos (c) vs. that for medical images (d).
Methodology
SAM2-3dMed Architecture
SAM2-3dMed builds on the SAM2 backbone, incorporating innovations to adapt to 3D data:
- SAM2 Backbone: Utilizes SAM2’s pre-trained Image Encoder for extracting features, processing slices independently.
- SRPP Module: Employs a self-supervised task to predict relative slice positions, enforcing the model to understand spatial context.
- BD Module: Enhances boundary segmentation via a parallel decoding branch with boundary-focused attention mechanisms.
These modules collectively optimize 3D segmentation, leveraging SAM2’s feature representations while adapting its mechanisms to capture volumetric dependencies.
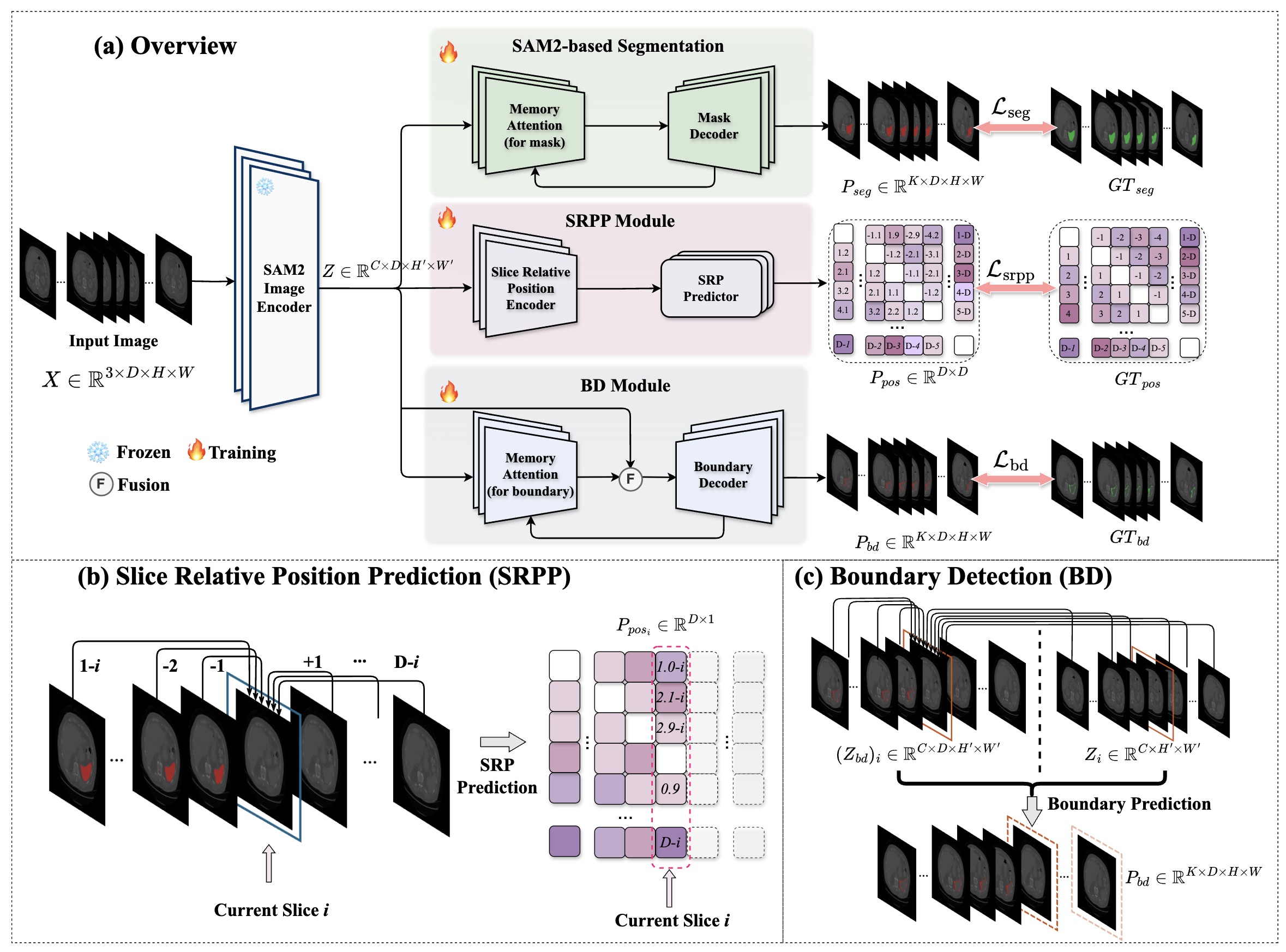
Figure 2: Overview of the proposed SAM2-3dMed Model.
Loss Function
The model employs a composite loss function:
Ltotal=Lseg+λ1Lsrpp+λ2Lbd
- Lseg: Dice loss for segmentation.
- Lsrpp: MSE loss for SRPP.
- Lbd: Weighted binary cross-entropy for boundary detection.
This structured loss approach fosters robust feature learning across tasks, improving volumetric segmentation accuracy.
Experiments
Datasets and Evaluation
The research utilized diverse datasets from the Medical Segmentation Decathlon (MSD), focusing on Lung, Spleen, and Pancreas tasks, to validate SAM2-3dMed against state-of-the-art baselines. Performance metrics included Dice, IoU, HD95, and NSD to measure segmentation overlap and boundary accuracy.
Results
SAM2-3dMed demonstrated superior performance across benchmarks, with notable Dice and NSD improvements, particularly in the Pancreas dataset. It significantly exceeded the capabilities of traditional CNN-based and newer Transformer-based models by enhancing boundary detection and spatial consistency:
Ablation Studies
Comprehensive ablation studies validated each module's contributions:
Conclusion
SAM2-3dMed represents a significant step towards adapting video segmentation models for 3D medical imaging. By focusing on inter-slice contextual understanding and boundary precision, it not only achieves state-of-the-art performance but also establishes a versatile framework for future adaptations of video-centric models to medical volumetric data, paving the way for enhanced clinical applications where data availability is limited.