- The paper introduces a novel automated discovery system that decontextualizes scientific data into keywords for exploring unexpected research connections.
- It outlines a multi-stage pipeline that transitions from graph-based inspiration to thesis reconstruction and rigorous evaluation using LLMs.
- Empirical results validate the approach with robust metrics like AUROC and embedding space analyses, demonstrating high alignment with expert research.
Spacer: Towards Engineered Scientific Inspiration
Introduction and Motivation
Spacer introduces a novel paradigm for automated scientific discovery, addressing the creative limitations inherent in transformer-based LLMs. The system is motivated by the observation that major scientific breakthroughs often arise from unexpected connections between disparate concepts, rather than incremental extensions of existing knowledge. Spacer operationalizes this insight through "deliberate decontextualization," decomposing scientific information into atomic units—keywords—and constructing new knowledge by exploring unexplored combinations within a large-scale keyword graph derived from 180,000 biological publications.
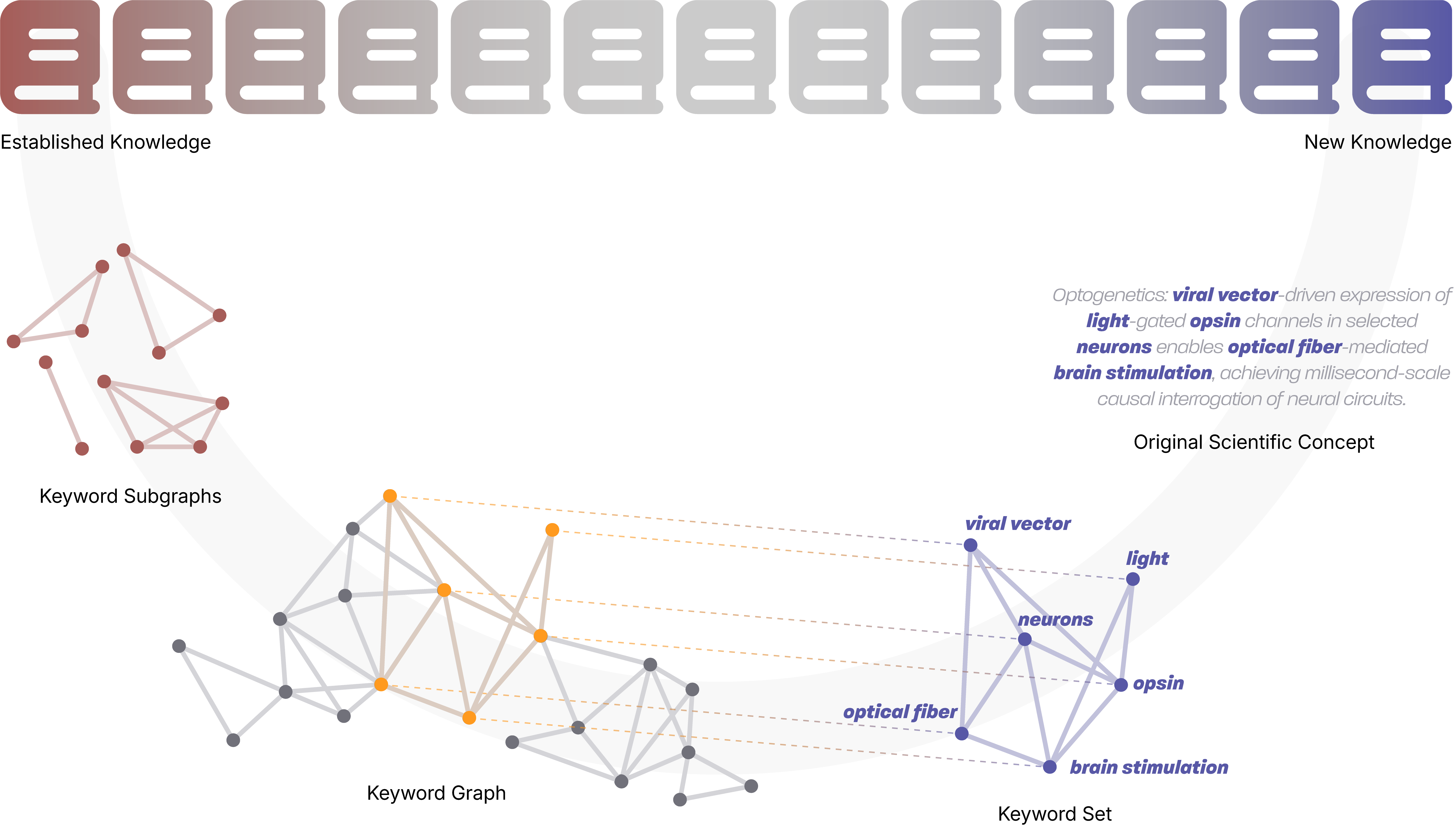
Figure 1: Schematic of Spacer's approach to engineered scientific inspiration.
System Architecture
Spacer is architected as a multi-stage pipeline, explicitly separating creative ideation from critical evaluation. The pipeline consists of four primary components:
- Nuri (Inspiration Engine): Graph-based algorithm for extracting high-potential keyword sets, leveraging edge weights computed from normalized logarithmic FWCI values to estimate the joint academic impact of keyword pairs.
- Revealing Framework: LLM-based module (Weaver and Sketcher) that transforms keyword sets into thesis paragraphs, reconstructing plausible research concepts and goals.
- Scaffolding Framework: Logic graph-based system that refines theses into structured Statements, supplementing them with validated rationales and evidence.
- Assessment Framework: Two-phase LLM-based evaluation (exploratory analysis and specified inspection) to ensure scientific plausibility and feasibility.
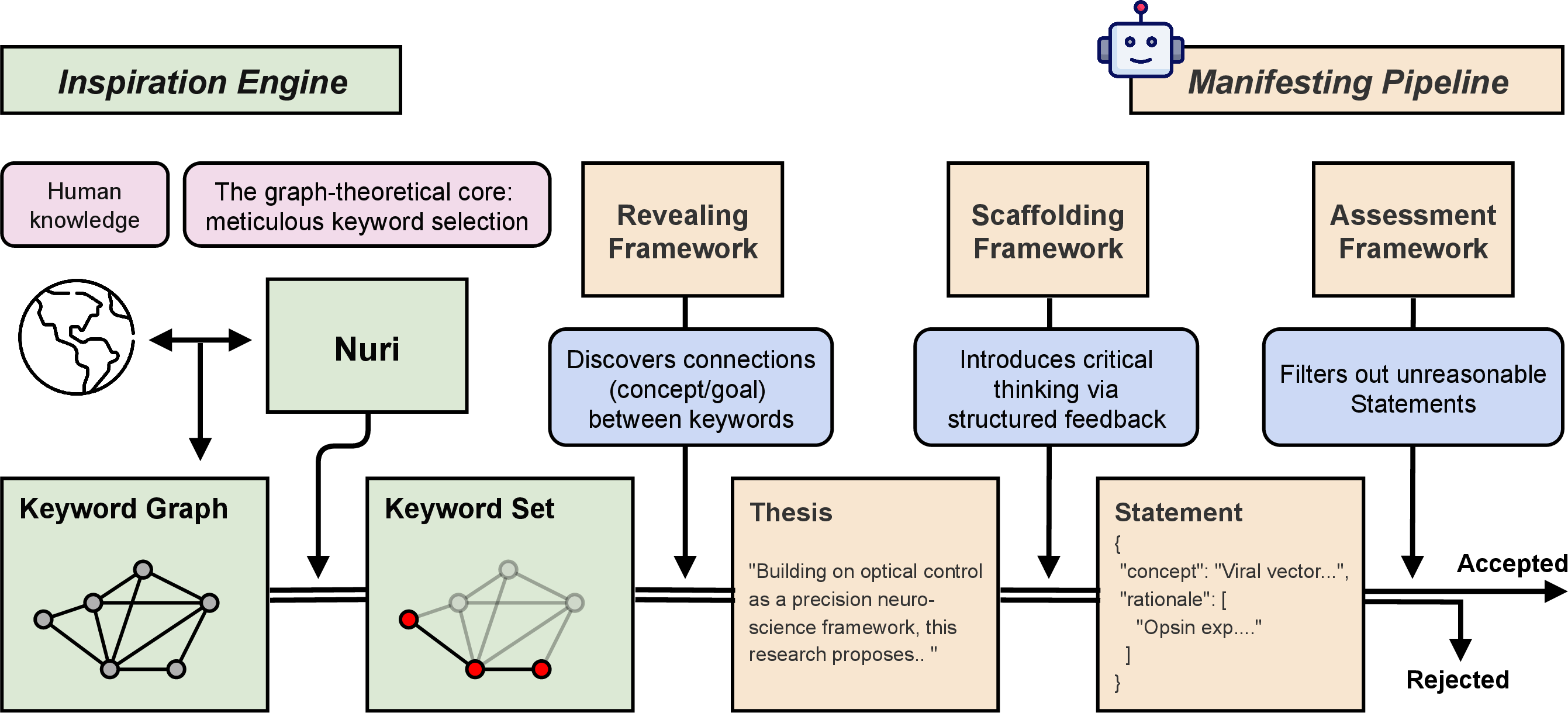
Figure 2: Architecture of Spacer.
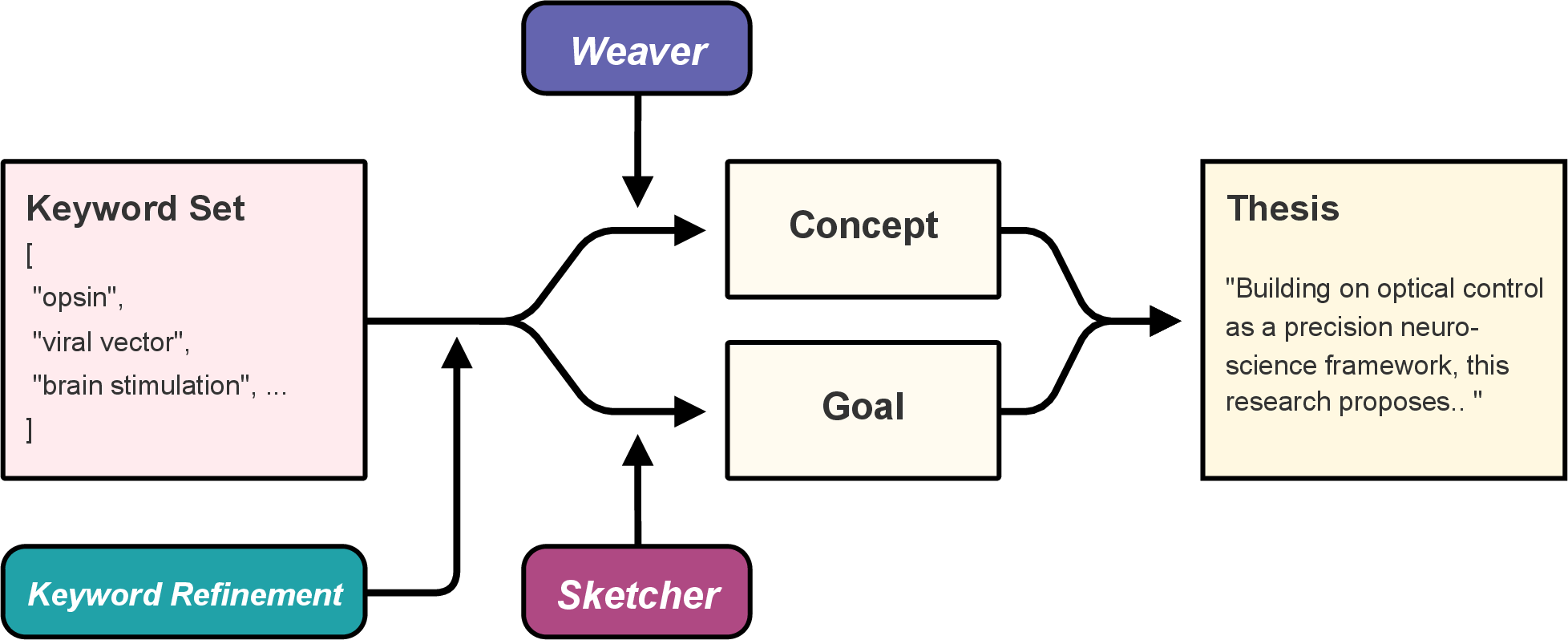
Figure 3: Schematic of the Revealing Framework.
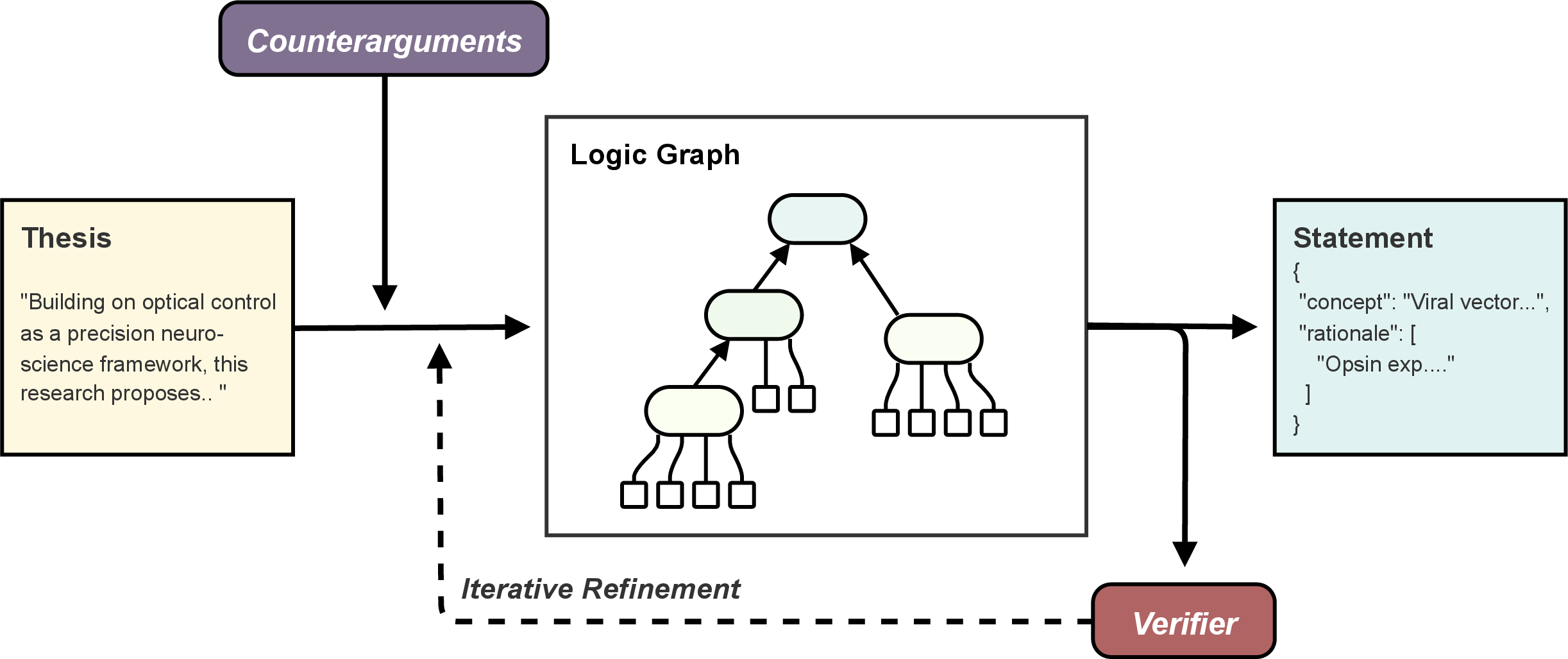
Figure 4: Schematic of the Scaffolding Framework.
Deliberate Decontextualization and Keyword Graph Construction
Spacer's core innovation is the deliberate removal of contextual information, reducing scientific knowledge to sets of keywords. This enables the system to circumvent the contextual bias and mode collapse typical of LLMs, which tend to reproduce established patterns from training data. Nuri constructs an undirected, weighted keyword graph G(P), where vertices are keywords and edge weights reflect the joint impact of keyword pairs across the corpus. The evaluation function fP assigns a normalized score to keyword sets, estimating their potential for high-impact research.
Empirical Results and Case Studies
Spacer demonstrates the ability to generate scientifically rigorous and novel concepts, as illustrated by three representative outputs:
- Restoring Calcium Oscillations in Hepatocellular Carcinoma: Proposes stochastic resonance via controlled noise injection to restore calcium signaling coherence, potentially suppressing malignant phenotypes.
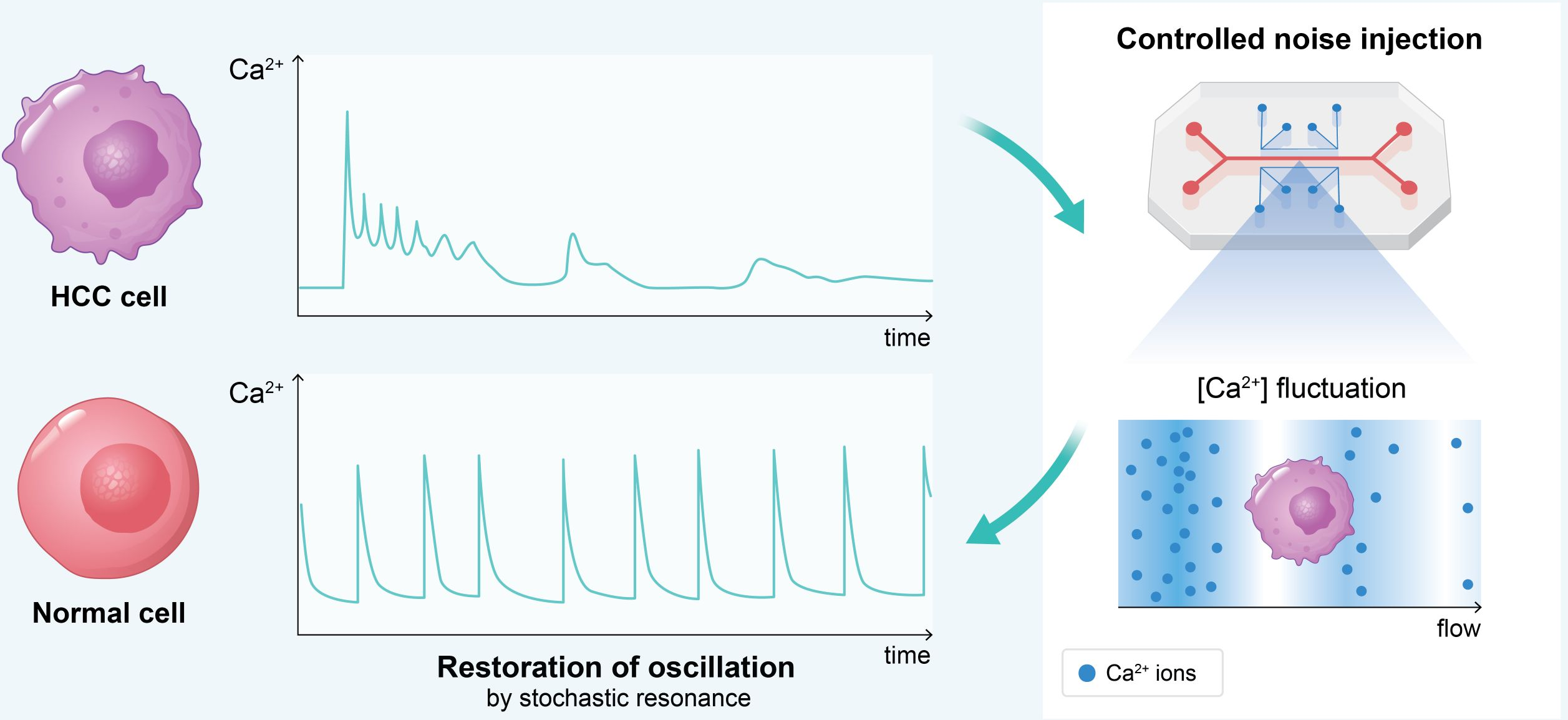
Figure 5: Hepatocellular carcinoma cells exhibit disrupted calcium oscillations. Controlled noise injection as extracellular calcium fluctuation could restore oscillatory coherence toward normal state, suppressing malignant phenotype.
- ATP Allocation Patterns Predict Cellular State Transitions: Suggests that quantifying ATP allocation across metabolic pathways enables prediction of cellular state transitions, integrating thermodynamic and kinetic modeling with experimental validation.
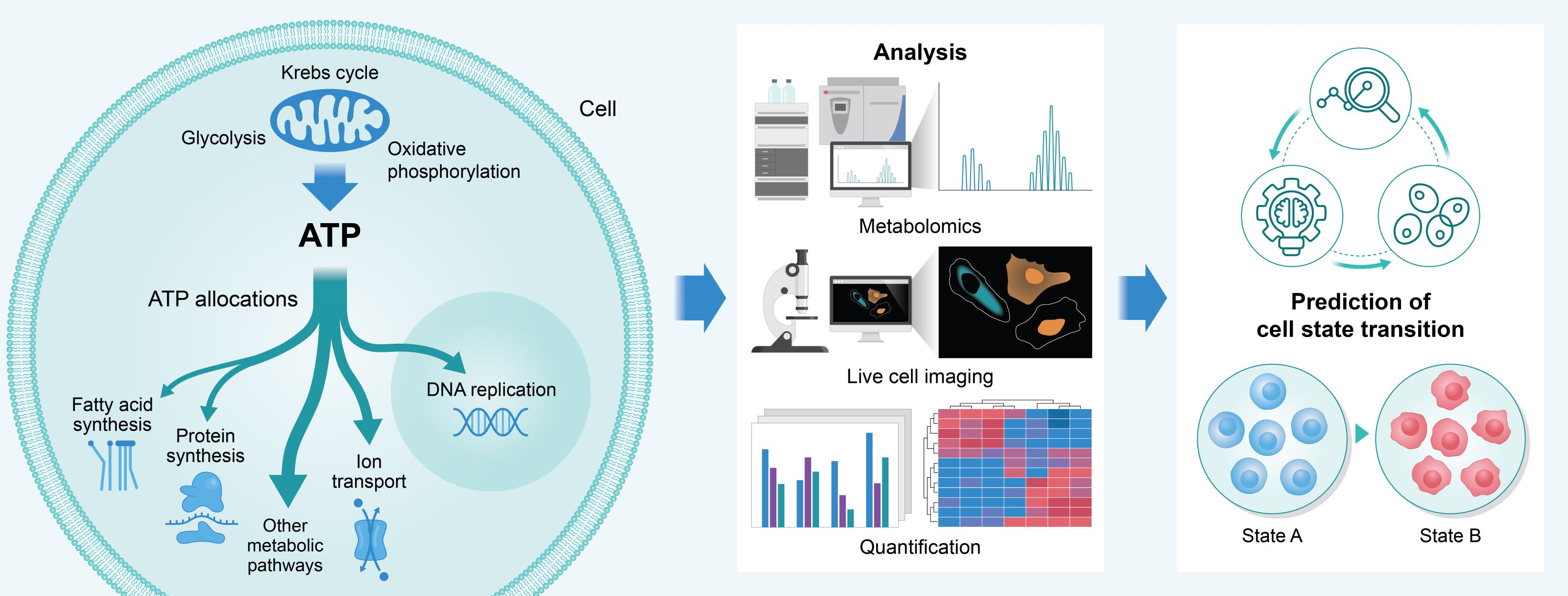
Figure 6: ATP is distributed across diverse metabolic pathways. Quantifying the allocation may enable prediction of cellular state transitions.
- Overexpressing Olfactory Receptors for Gut Microbiome Control: Argues for engineering intestinal epithelial cells to overexpress olfactory receptors, enabling localized antimicrobial peptide secretion in response to microbial metabolites.
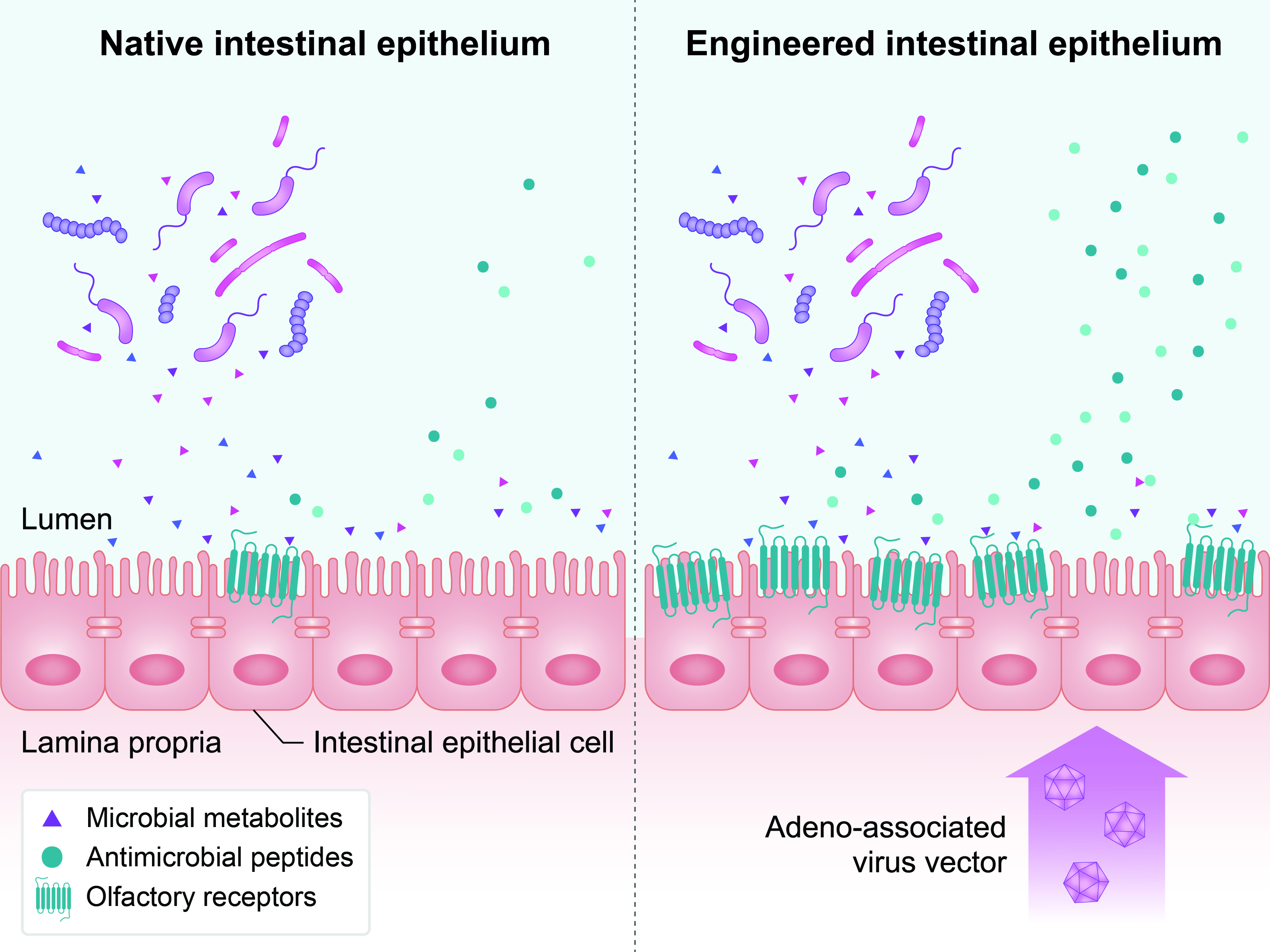
Figure 7: Intestinal epithelial cells can be engineered to overexpress olfactory receptors. This may lead to localized antimicrobial peptide secretion, enhancing intestinal microbial regulation.
Validation and Quantitative Evaluation
Nuri's Impact Estimation
Nuri's evaluation function achieves an AUROC of 0.737±0.025 in classifying high-impact versus low-impact papers, indicating substantial discriminative power. The distribution of FWCI values for papers with high EVAL scores is strongly shifted toward high-impact, and the function robustly distinguishes paper-derived keyword sets from random sets (AUC 0.996±0.003).
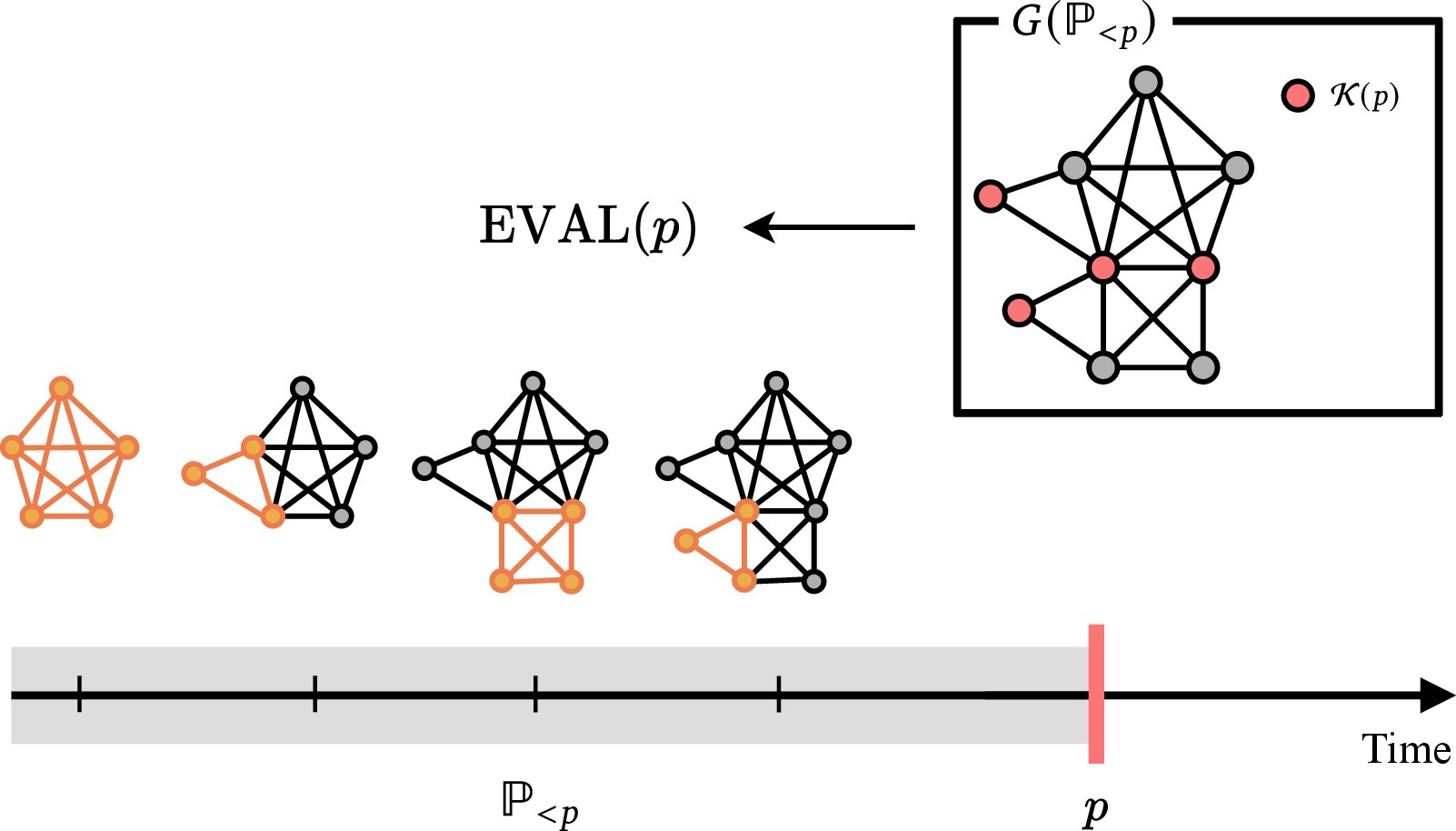
Figure 8: Schematic of the evaluation process of Nuri. For each paper, a complete graph is constructed with its keywords as vertices.
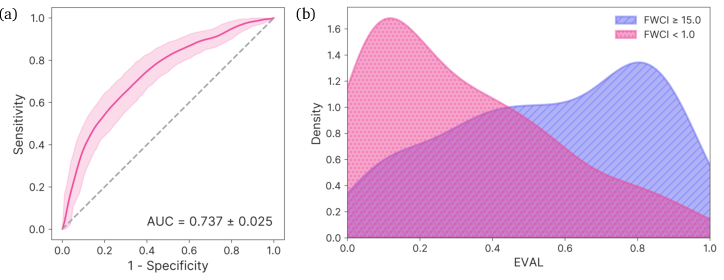
Figure 9: Performance of EVAL on a validation set of 200 high-impact and 200 low-impact papers. AUROC = 0.737±0.025.
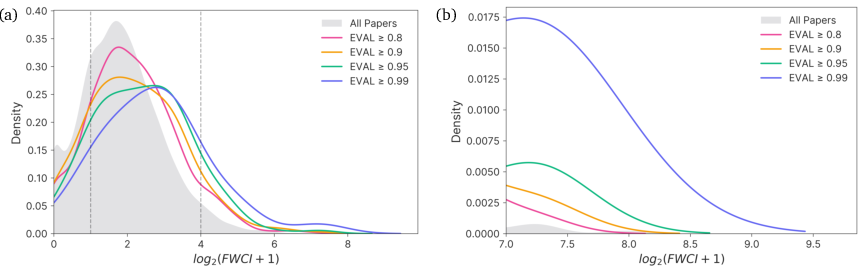
Figure 10: Distribution of log2(FWCI+1) for 10,000 papers, stratified by EVAL threshold.
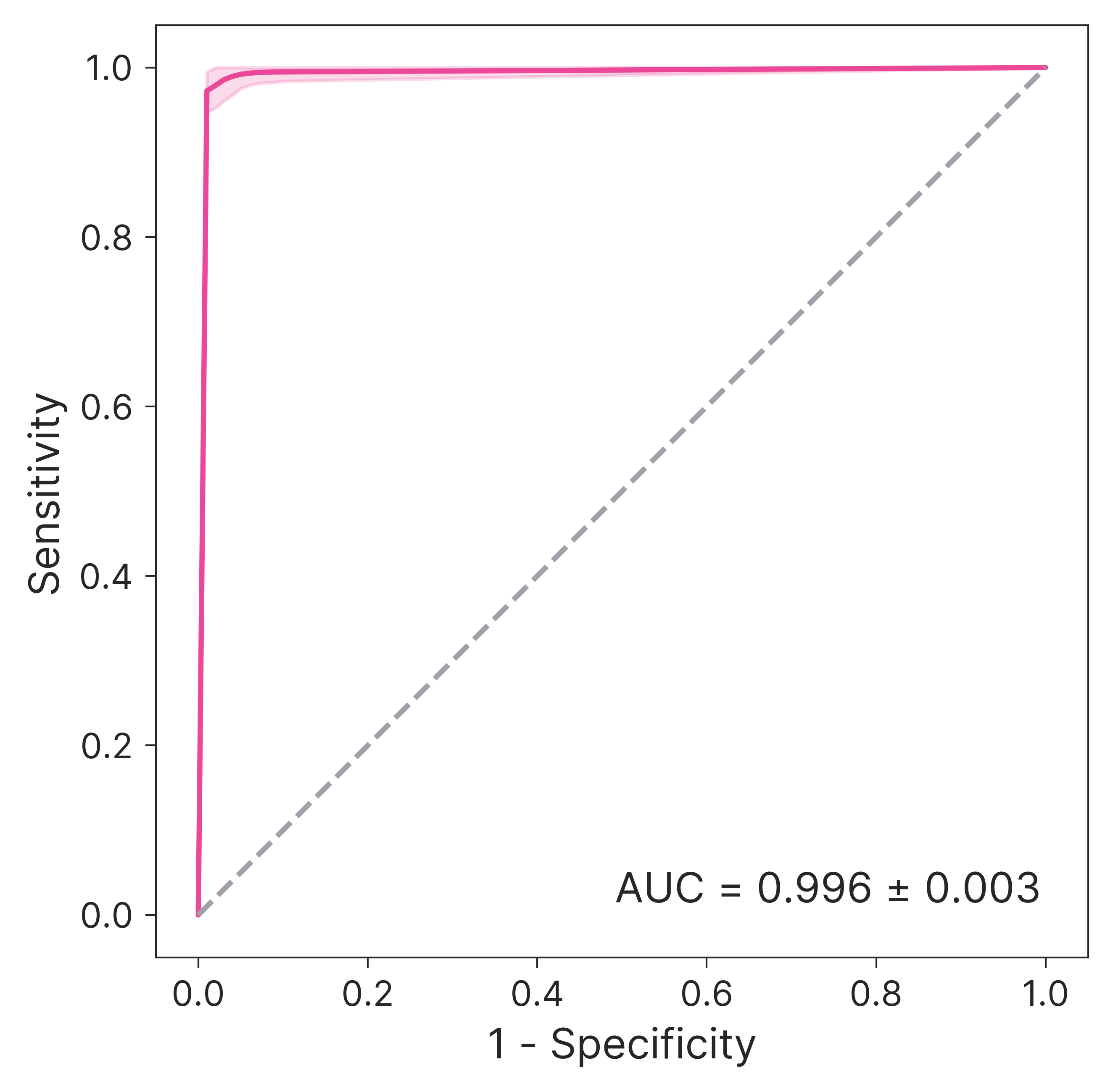
Figure 11: ROC curve of EVAL for distinguishing paper-derived from random keyword sets.
Thesis Reconstruction and Semantic Alignment
Weaver reconstructs thesis paragraphs from keyword sets with high fidelity: over 85% of reconstructed theses are judged as logically and semantically equivalent to original human-authored research, across multiple journals and evaluation criteria.
Embedding Space Analysis
Spacer's outputs are significantly closer to published human research than those of SOTA LLMs, as measured by PCA, LDA, and energy distance metrics in a 364-sample embedding analysis. Spacer exhibits minimal semantic drift and high overlap with expert research, while LLM-generated ideas display greater variance and lower alignment.
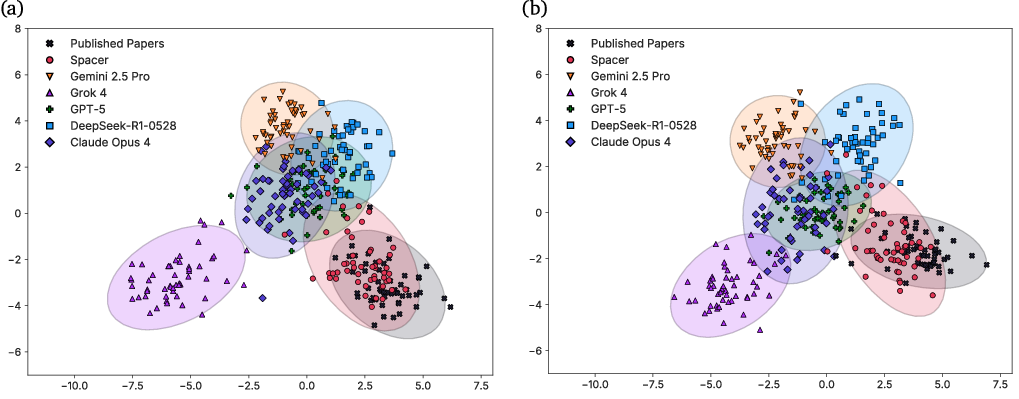
Figure 12: LDA results of 364 research theses from Spacer, 5 SOTA LLMs, and ideations of published papers.
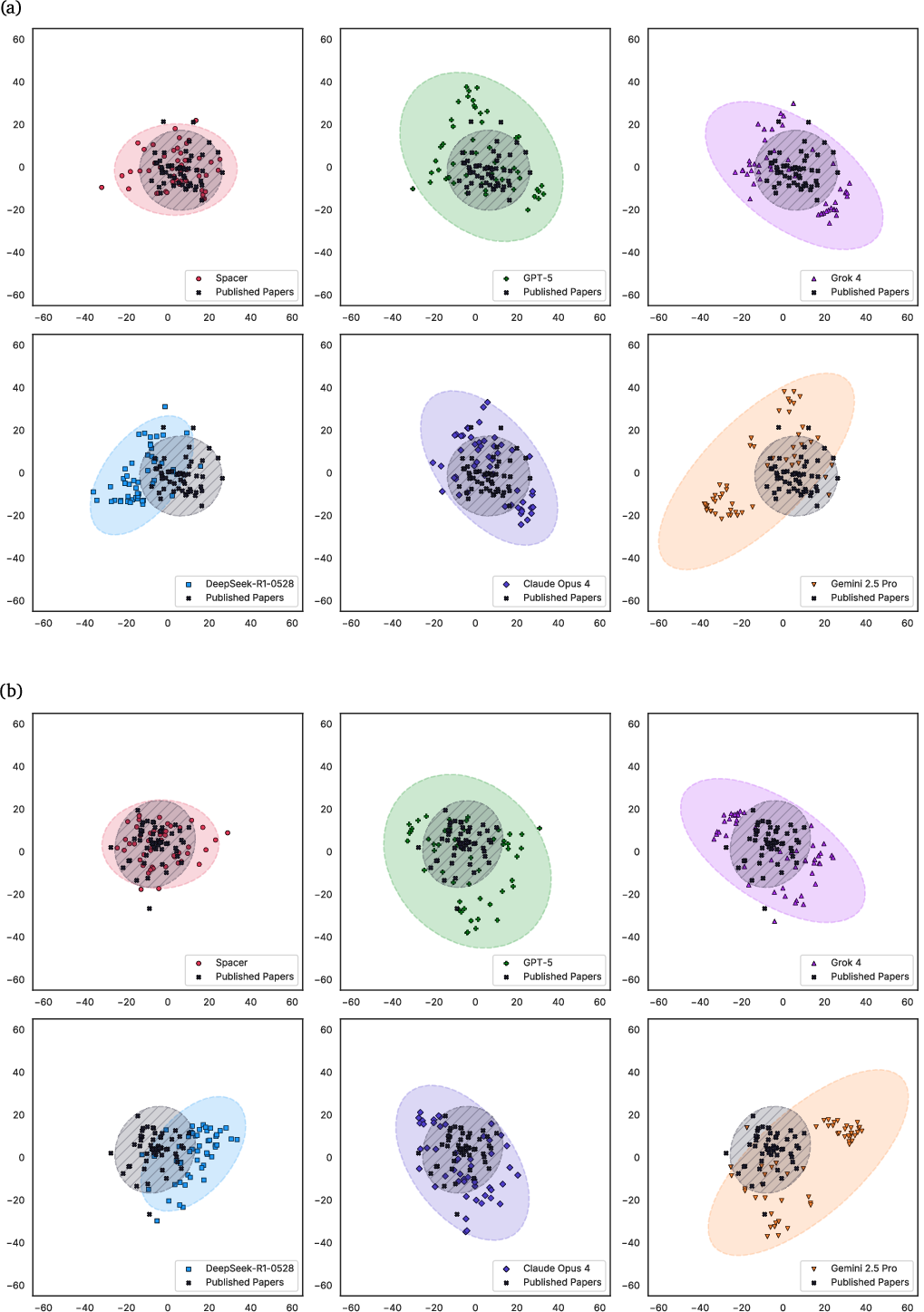
Figure 13: PCA results of 364 research theses from Spacer, 5 SOTA LLMs, and ideations of published papers.
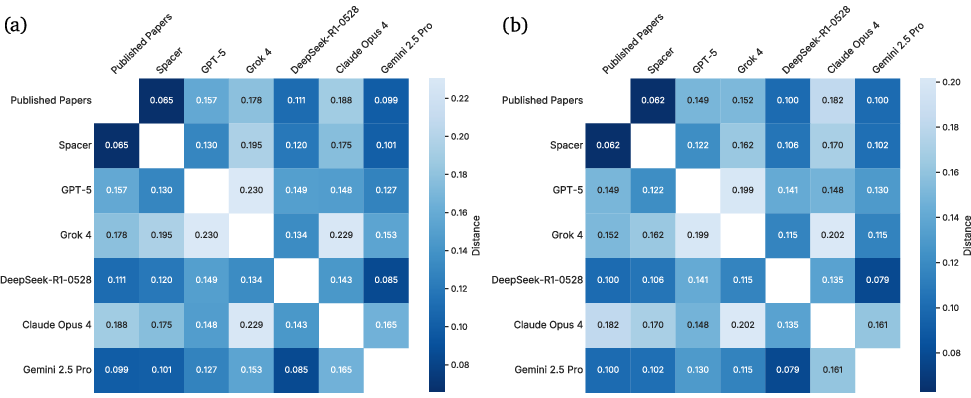
Figure 14: Heatmap of energy distances between Spacer, 5 SOTA LLMs, and ideations of published papers.
Technical Implementation
Spacer is implemented in Rust for type safety and parallelism, with a custom agentic AI framework. LLM inference is the primary computational bottleneck, but the system is agnostic to model architecture and can integrate future advances in language modeling. Fine-tuning is performed on open-weight models (DeepSeek-R1, Gemma 3), and proprietary SOTA LLMs (o3, Grok 4, Gemini 2.5 Pro, Claude Opus 4) are used for backbone components. Training and inference are conducted on 8×NVIDIA H100 GPUs and high-memory CPU nodes.
Limitations and Future Directions
Spacer's approach is optimal for LLM-driven scientific ideation but does not claim to exhaust all sources of human inspiration. The system can be further augmented by improved base models, reinforcement learning, and non-autoregressive architectures. Extension to other scientific domains is straightforward, and future work will focus on automating the materialization of research plans and experimental validation, leveraging robotics and in silico tools.
Conclusion
Spacer represents a rigorous framework for engineered scientific inspiration, overcoming the creative limitations of LLMs through deliberate decontextualization and graph-based keyword set exploration. Empirical validation demonstrates Spacer's capacity to generate original, scientifically plausible concepts with semantic alignment to expert research. The system provides a scalable, cost-effective platform for automated knowledge expansion, with broad applicability across scientific domains. Spacer marks a substantive advance toward the automation of scientific discovery and the realization of artificial superintelligence.













