An AI system to help scientists write expert-level empirical software (2509.06503v1)
Abstract: The cycle of scientific discovery is frequently bottlenecked by the slow, manual creation of software to support computational experiments. To address this, we present an AI system that creates expert-level scientific software whose goal is to maximize a quality metric. The system uses a LLM and Tree Search (TS) to systematically improve the quality metric and intelligently navigate the large space of possible solutions. The system achieves expert-level results when it explores and integrates complex research ideas from external sources. The effectiveness of tree search is demonstrated across a wide range of benchmarks. In bioinformatics, it discovered 40 novel methods for single-cell data analysis that outperformed the top human-developed methods on a public leaderboard. In epidemiology, it generated 14 models that outperformed the CDC ensemble and all other individual models for forecasting COVID-19 hospitalizations. Our method also produced state-of-the-art software for geospatial analysis, neural activity prediction in zebrafish, time series forecasting and numerical solution of integrals. By devising and implementing novel solutions to diverse tasks, the system represents a significant step towards accelerating scientific progress.
Collections
Sign up for free to add this paper to one or more collections.
Summary
- The paper introduces an AI system that leverages LLM-based code rewriting within a tree search to iteratively generate and refine expert-level empirical software.
- It achieves superhuman performance by recombining existing methods and integrating external research, outperforming benchmarks in genomics, epidemiology, geospatial analysis, and more.
- The system's versatile methodology and scalable design offer new avenues for automated scientific software discovery and optimization.
AI-Driven Tree Search for Expert-Level Empirical Software in Science
Introduction
The paper "An AI system to help scientists write expert-level empirical software" (2509.06503) presents a general-purpose AI system that automates the creation and optimization of empirical scientific software. The system leverages a LLM as a code rewriter, embedded within a Tree Search (TS) framework, to iteratively generate, evaluate, and refine code solutions for scorable scientific tasks. The approach is benchmarked across diverse domains, including genomics, epidemiology, geospatial analysis, neuroscience, time series forecasting, and numerical analysis, demonstrating expert-level or superhuman performance on public leaderboards and academic benchmarks.
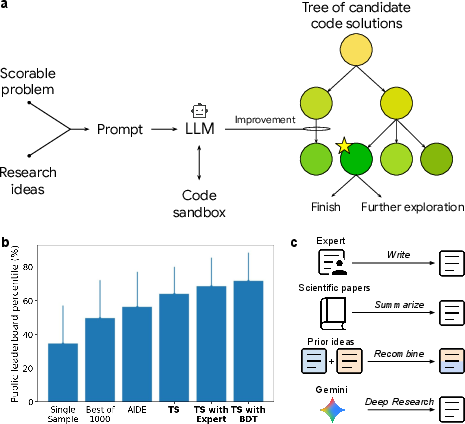
Figure 1: Schematic and performance of the method, showing the LLM-driven code mutation system embedded in a tree search, and its performance on Kaggle Playground benchmarks.
System Architecture and Methodology
The core system is a code mutation engine that uses an LLM to propose code modifications, guided by a TS algorithm that balances exploration and exploitation. The process is as follows:
- Task Definition: The user specifies a scorable task, i.e., a problem with a well-defined, machine-evaluable quality metric.
- Prompt Construction: The LLM is prompted with the task description, evaluation metric, relevant data, and optionally, research ideas or method summaries.
- Code Generation and Evaluation: The LLM generates candidate code, which is executed in a sandbox and scored.
- Tree Search: The TS algorithm (PUCT variant) maintains a tree of candidate solutions, selecting nodes for expansion based on a combination of empirical score and exploration bonus.
- Research Idea Injection: The system can incorporate external research ideas, either manually provided or automatically retrieved from literature, to guide code generation.
- Recombination: The system can synthesize hybrid solutions by recombining features from multiple base methods.
This approach generalizes and extends prior work in genetic programming, generative programming, LLM-based code generation, and AutoML, but is distinguished by its semantic-aware code mutation, integration of external research, and generality across scientific domains.
Empirical Results Across Scientific Domains
Genomics: Single-Cell RNA-seq Batch Integration
The system was evaluated on the OpenProblems v2.0.0 benchmark for scRNA-seq batch integration, a task requiring removal of batch effects while preserving biological variability. The TS-based system, when seeded with method summaries from the literature, consistently outperformed published methods, including a 14% improvement over the best prior method (ComBat) using a novel recombination of ComBat and BBKNN.
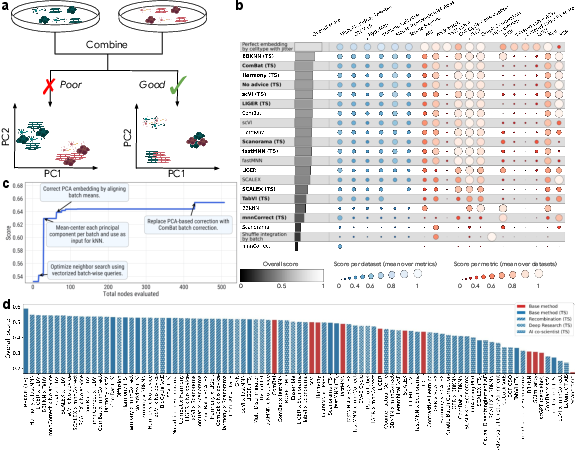
Figure 2: Performance of tree search on scRNA-seq batch integration, showing superior overall scores compared to published methods and the impact of code innovations.
Systematic recombination of method ideas yielded 24/55 hybrids that outperformed both parent methods, and 40/87 generated methods exceeded the best published leaderboard results. Embedding-based clustering of generated code revealed the system's ability to explore diverse algorithmic spaces.
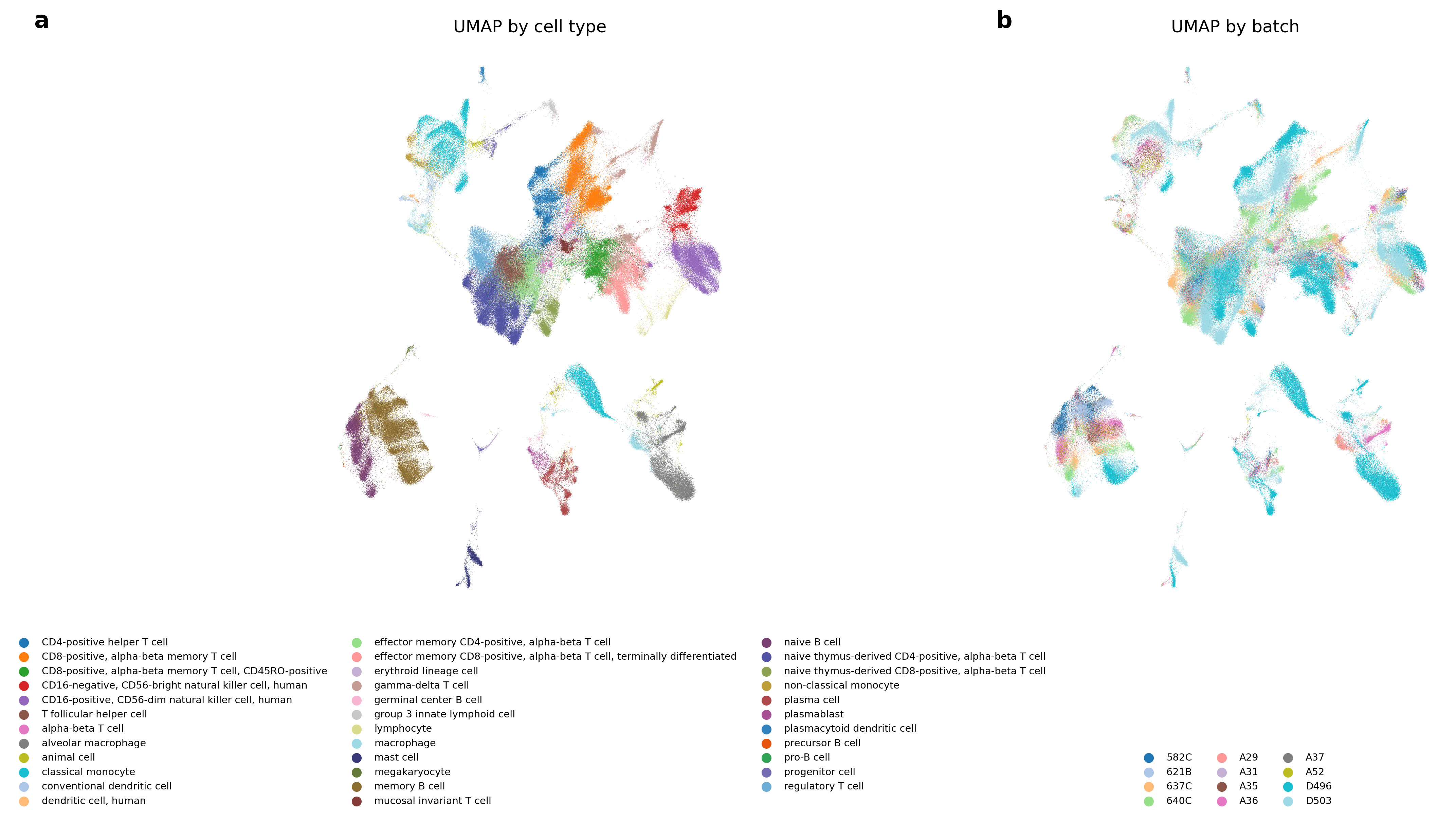
Figure 3: UMAP projection of BBKNN (TS) embeddings on the Immune Cell Atlas, showing effective batch mixing and cell-type separation.
Epidemiology: COVID-19 Hospitalization Forecasting
On the CDC COVID-19 Forecast Hub, the system generated 14 models that outperformed the official ensemble and all individual models for forecasting hospitalizations, as measured by Weighted Interval Score (WIS). The system replicated, recombined, and innovated upon existing models, with hybrid models consistently outperforming their parents. Notably, the system discovered that combining climatological baselines with autoregressive or machine learning models yielded robust, high-performing forecasts.
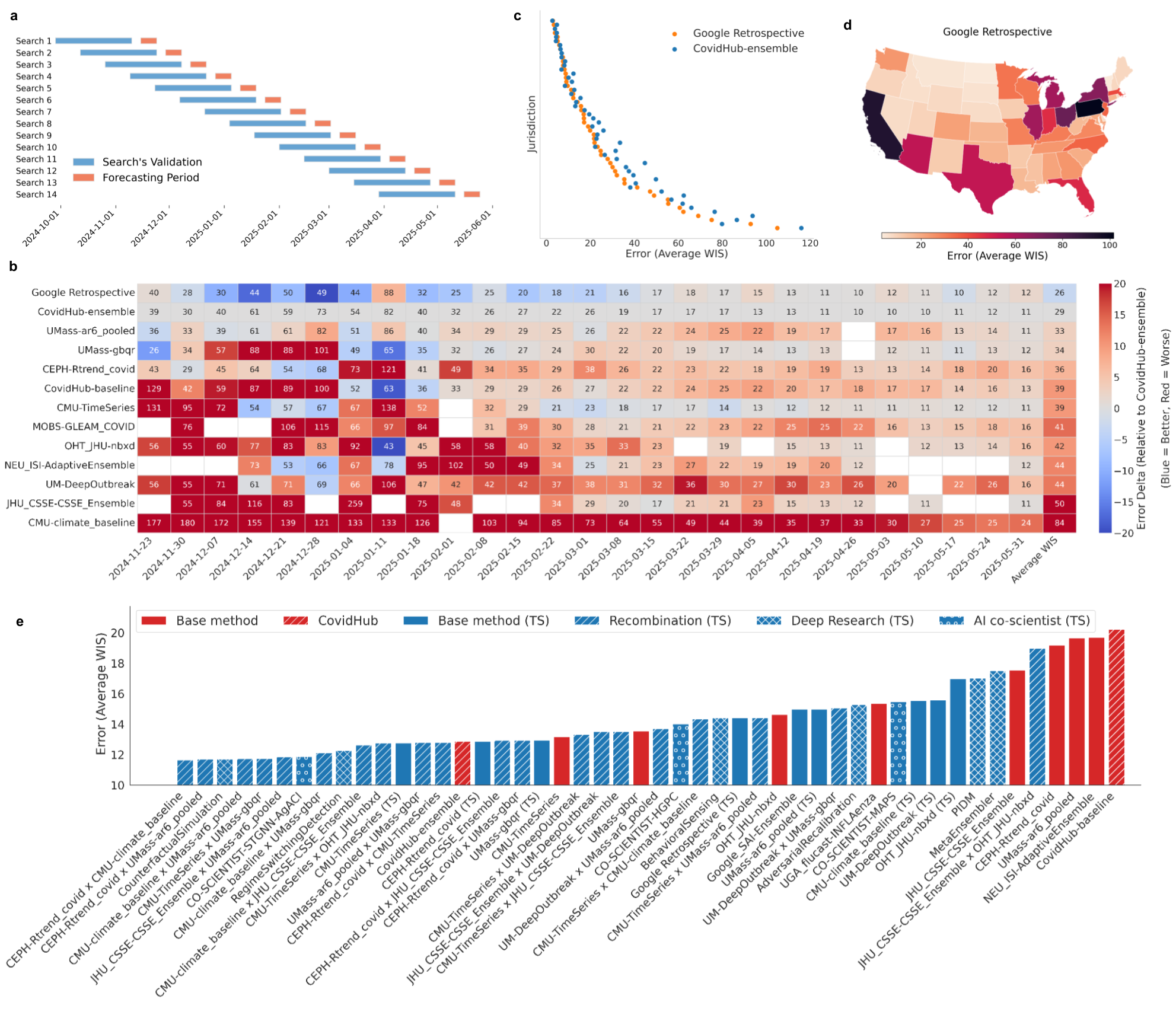
Figure 4: COVID-19 forecasting performance, with the system's models outperforming the CDC ensemble across jurisdictions and time periods.
Geospatial Analysis: Remote Sensing Image Segmentation
For semantic segmentation of satellite imagery (DLRSD), the system produced solutions based on U-Net++ and SegFormer architectures, leveraging advanced encoders and test-time augmentation. All top solutions exceeded the best published results, achieving mIoU > 0.80.
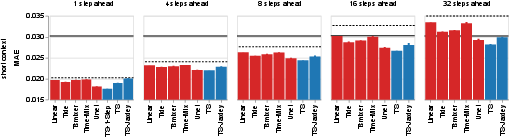
Figure 5: Comparison of tree search solutions to baselines on ZAPBench, showing lower MAE (better performance) for the system's models.
Neuroscience: Whole-Brain Neural Activity Prediction
On ZAPBench, the system generated time-series models that outperformed both linear and deep learning baselines, and approached the performance of computationally intensive video-based models, but with orders-of-magnitude lower resource requirements. The system also demonstrated the ability to integrate biophysical neuron simulators (Jaxley) into performant hybrid models.
Time Series Forecasting: GIFT-Eval
The system achieved state-of-the-art results on the GIFT-Eval benchmark, both with per-dataset and unified solutions. The unified model, discovered via tree search, sequentially decomposes time series into base, trend, seasonality, datetime, and residual components, and is highly competitive with foundation models and deep learning approaches.
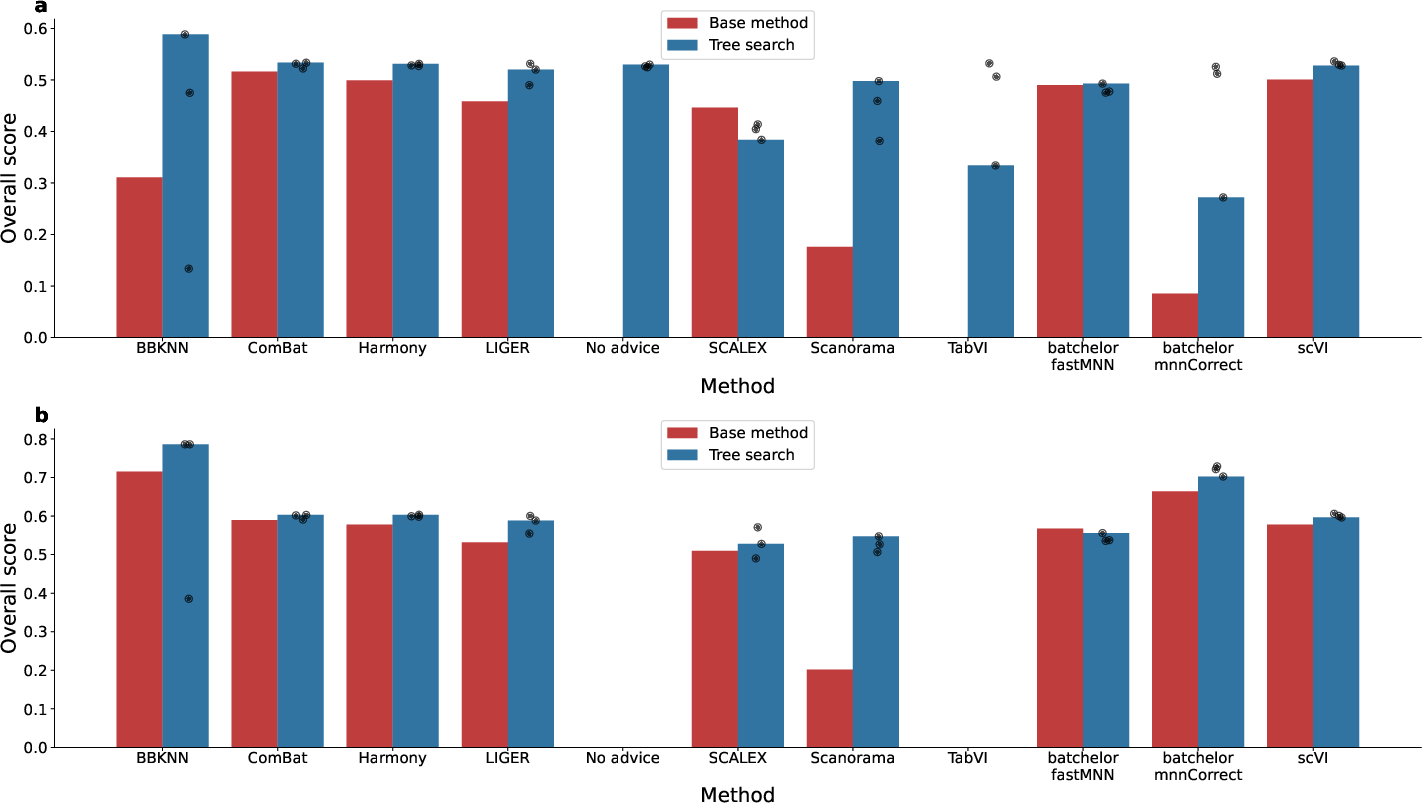
Figure 6: Distribution of solution categories on GIFT-Eval, showing the prevalence of ensemble and gradient boosting methods among top solutions.
Numerical Analysis: Difficult Integrals
The system evolved a numerical integration routine that outperformed scipy.integrate.quad() on a set of challenging oscillatory integrals, correctly solving 17/19 held-out cases where the standard library failed. The solution combined adaptive domain partitioning with Euler's series acceleration, and is suitable as a drop-in replacement for quad() in pathological cases.
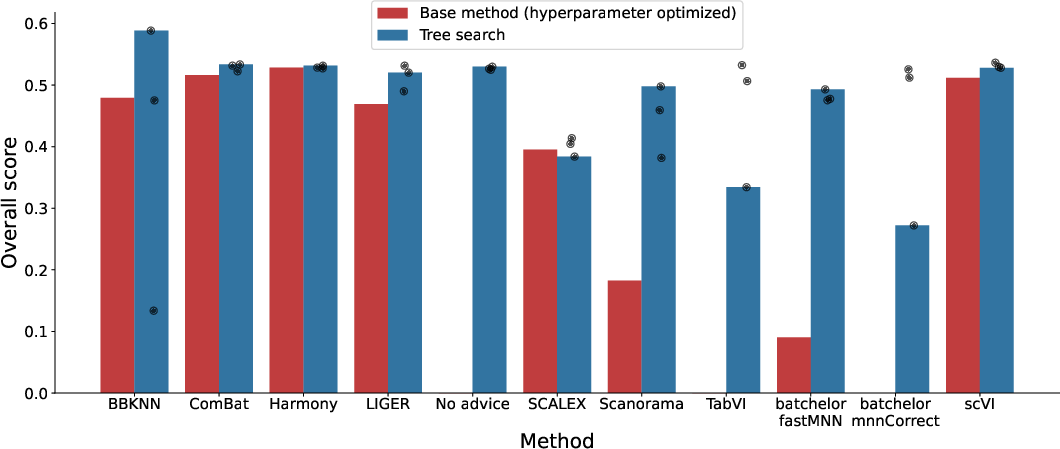
Figure 7: Scores of the best numerical integration routine on held-out integrals, with the system's method solving nearly all cases to high accuracy.
Implementation Details and Trade-offs
- Tree Search: The PUCT-based TS is adapted for non-enumerable, stochastic LLM outputs. Node selection is based on rank scores, and expansion is driven by LLM sampling.
- LLM Prompting: Prompts are dynamically constructed to include task context, code history, and research ideas. Injection of method summaries or literature-derived ideas is critical for expert-level performance.
- Evaluation: All code is executed in a sandboxed environment, with domain-specific metrics (e.g., mIoU, WIS, MASE) used for scoring.
- Resource Requirements: Typical searches involve hundreds to thousands of nodes, with per-task runtimes ranging from hours (single GPU) to days (large-scale benchmarks). The approach is highly parallelizable.
- Reproducibility: The system's outputs, including code and search trees, are open-sourced for inspection and reuse.
Theoretical and Practical Implications
The work demonstrates that LLM-driven tree search, augmented with external research and recombination, can systematically discover, optimize, and hybridize expert-level scientific software across domains. The approach generalizes beyond AutoML by encompassing arbitrary empirical software, including pre-processing, simulation, and algorithmic heuristics. The system's ability to rapidly explore vast solution spaces, integrate literature, and synthesize novel hybrids has direct implications for accelerating scientific discovery in any field where tasks are scorable.
Theoretically, the results suggest that the combination of semantic-aware code mutation (via LLMs) and structured search (via TS) can overcome the limitations of both random mutation (genetic programming) and one-shot code generation. The system's performance on public leaderboards and academic benchmarks provides strong evidence for the viability of automated empirical software discovery.
Future Directions
- Automated Literature Mining: Further automation of research idea extraction and injection, leveraging retrieval-augmented LLMs and scientific knowledge graphs.
- Multi-Agent Collaboration: Integration with multi-agent systems for distributed exploration and cross-domain synthesis.
- Interpretability and Verification: Enhanced tools for code verification, interpretability, and human-in-the-loop refinement.
- Scaling and Efficiency: Optimization of search strategies for larger codebases and more complex tasks, including distributed and asynchronous search.
- Generalization to Unscorable Tasks: Extension to tasks where evaluation is not strictly quantitative, via surrogate metrics or human feedback.
Conclusion
This work establishes a new paradigm for automated scientific software creation, demonstrating that LLM-driven tree search, guided by research ideas and recombination, can achieve and surpass expert-level performance across a wide range of empirical scientific tasks. The system's generality, extensibility, and empirical success position it as a foundational tool for accelerating computational science, with broad implications for the future of AI-assisted research and discovery.
Paper Prompts
Sign up for free to create and run prompts on this paper using GPT-5.
Follow-up Questions
- How does the tree search algorithm balance exploration and exploitation in the code generation process?
- What specific role does LLM-driven code rewriting play in enhancing expert-level performance?
- How is external research integrated into the system to guide method recombination?
- What benchmarks and metrics demonstrate the system's superiority across various scientific domains?
- Find recent papers about automated scientific software discovery.
Related Papers
- The AI Scientist: Towards Fully Automated Open-Ended Scientific Discovery (2024)
- Transforming Science with Large Language Models: A Survey on AI-assisted Scientific Discovery, Experimentation, Content Generation, and Evaluation (2025)
- The AI Scientist-v2: Workshop-Level Automated Scientific Discovery via Agentic Tree Search (2025)
- IRIS: Interactive Research Ideation System for Accelerating Scientific Discovery (2025)
- From Automation to Autonomy: A Survey on Large Language Models in Scientific Discovery (2025)
- InternAgent: When Agent Becomes the Scientist -- Building Closed-Loop System from Hypothesis to Verification (2025)
- AI-Researcher: Autonomous Scientific Innovation (2025)
- Open Source Planning & Control System with Language Agents for Autonomous Scientific Discovery (2025)
- AlphaGo Moment for Model Architecture Discovery (2025)
- How Far Are AI Scientists from Changing the World? (2025)
Authors (42)
Tweets
YouTube
HackerNews
- An AI system to help scientists write expert-level empirical software (8 points, 1 comment)
- An AI system to help scientists write expert-level empirical software (3 points, 1 comment)
- An AI system to help scientists write expert-level empirical software (107 points, 13 comments)
- An AI system to help scientists write expert-level empirical software (led by Google DeepMind) (29 points, 6 comments)
- Google + MIT + Harvard just created an AI system that can generate scientific software that can model and outperform the best human tools and models across scientific disciplines!!! (28 points, 1 comment)
- An AI system to help scientists write expert-level empirical software, Aygün et al. 2025 (3 points, 0 comments)
- An AI system to help scientists write expert-level empirical software (0 points, 2 comments)
alphaXiv
- An AI system to help scientists write expert-level empirical software (67 likes, 3 questions)