- The paper introduces Kvasir-VQA-x1, a dataset that adds 159,549 robust QA pairs to improve clinical reasoning in GI endoscopy.
- It employs structured QA generation and weak augmentation techniques to simulate imaging artifacts and capture reasoning complexity.
- Fine-tuned vision-language models demonstrate enhanced performance and robustness, highlighting the dataset's potential to advance clinical AI.
Kvasir-VQA-x1: A Multimodal Dataset for Medical Reasoning
The paper introduces Kvasir-VQA-x1, a novel large-scale dataset designed to advance medical visual question answering (MedVQA) in gastrointestinal (GI) endoscopy. This dataset expands upon the original Kvasir-VQA by incorporating 159,549 new question-answer pairs, crafted to evaluate deeper clinical reasoning and model robustness through visual augmentations that mimic common imaging artifacts. The dataset supports two evaluation tracks: standard VQA performance and robustness against visual perturbations, aiming to accelerate the development of reliable multimodal AI systems for clinical use.
Background and Motivation
MedVQA aims to develop systems capable of interpreting medical images and answering clinically relevant questions, holding transformative potential for enhancing diagnostic accuracy and improving patient care. Unlike general-domain VQA, MedVQA presents unique challenges due to the complexity of medical images and the domain-specific knowledge required. The shift towards generative models in MedVQA, driven by advancements in LLMs and VLMs, is hindered by the lack of complex, domain-aligned datasets. GI endoscopy, despite its visual complexity and clinical content, has received limited attention in VQA research. Existing GI-specific resources often feature QA pairs centered on simple tasks, failing to capture the reasoning depth required for advanced clinical understanding. Kvasir-VQA-x1 addresses these limitations by providing a reasoning-intensive dataset with linguistic diversity, visual robustness, and complexity scoring.
Dataset Construction and Features
Kvasir-VQA-x1 builds upon the original Kvasir-VQA dataset, which includes 6,500 GI endoscopic images paired with 58,849 QA pairs. The construction of Kvasir-VQA-x1 involved two major enhancements: the generation of complex question-answer pairs and weak image augmentation for enhanced robustness. To promote reasoning beyond simple recall, the authors employed a structured pipeline involving QA grouping, combinatorial sampling, and prompt engineering using Qwen3-30B-A3B LLM. Each new QA pair includes an image (original or augmented), a complex question, a naturalized answer, the JSON-encoded original QA(s), and a complexity score from 1 to 3. To account for minor variations in imaging, 10 weakly augmented versions were generated for each original image using random resized crops, random rotations, random affine transformations, and color jitter.
Dataset Statistics and Evaluation Tracks
The final Kvasir-VQA-x1 dataset comprises 159,549 QA pairs. The dataset is released with only the original images, and scripts are provided to generate weakly augmented versions. Two evaluation settings are defined: the original setting (QA pairs referencing only the original images) and the transformed setting (QA pairs referencing weakly augmented images). This dual-track framework allows for transparent comparison across models while surfacing failure modes that traditional benchmarks may obscure.
Experimental Setup and Results
The authors fine-tuned two prominent vision-LLMs, MedGemma and Qwen2.5-VL, using LoRA. For Qwen2.5-VL, two fine-tuning variants were explored: one using the original image set and another using a transformed image set. Performance was evaluated using two distinct datasets (normal and transformed) and a comprehensive suite of standard VQA and NLP metrics, including ROUGE-1, ROUGE-2, ROUGE-L, METEOR, CHRF++, BLEU, BLEURT, and BERT-F1. Evaluation was performed at multiple granularities, including intermediate checkpoint evaluation, overall performance, categorical evaluation, and complexity-based evaluation. An automated evaluation protocol using a powerful LLM as a structured adjudicator was implemented to address the limitations of traditional n-gram-based metrics in capturing clinical accuracy and semantic correctness.
Key Findings and Implications
The fine-tuned models demonstrated a dramatic leap in performance, highlighting the transformative impact of domain-specific fine-tuning. The architectural superiority and scale become the deciding factors at the performance ceiling. Qwen's flexible image resolution and hierarchical vision features might provide richer spatial cues, contributing to its stronger performance on localization-dependent tasks. Incorporating visual augmentations during fine-tuning effectively improved model robustness. Models trained on transformed images demonstrated strong invariance to input perturbations, maintaining stable performance across both validation domains. Augmentation-informed training can be instrumental in reducing performance variance and ensuring consistent outputs across heterogeneous input conditions.
Limitations and Future Work
Limitations include dataset specificity, evaluation protocol, and persistent error modes. Future directions include advanced training strategies, explicit spatial and metric supervision, data augmentation, and refined evaluation. Homogeneity bias in the LLM-as-a-judge evaluation framework can be addressed by incorporating adjudication using structurally distinct LLMs.
Conclusion
Kvasir-VQA-x1 is a comprehensive VQA dataset designed to advance the development of multimodal AI systems in GI endoscopy. The dataset addresses key limitations of existing MedVQA datasets by increasing linguistic and reasoning diversity and provides a clear framework for assessing the inferential capabilities of AI models. The authors hope to foster a collaborative effort towards building more trustworthy and clinically impactful AI in gastroenterology and other medical specialties.
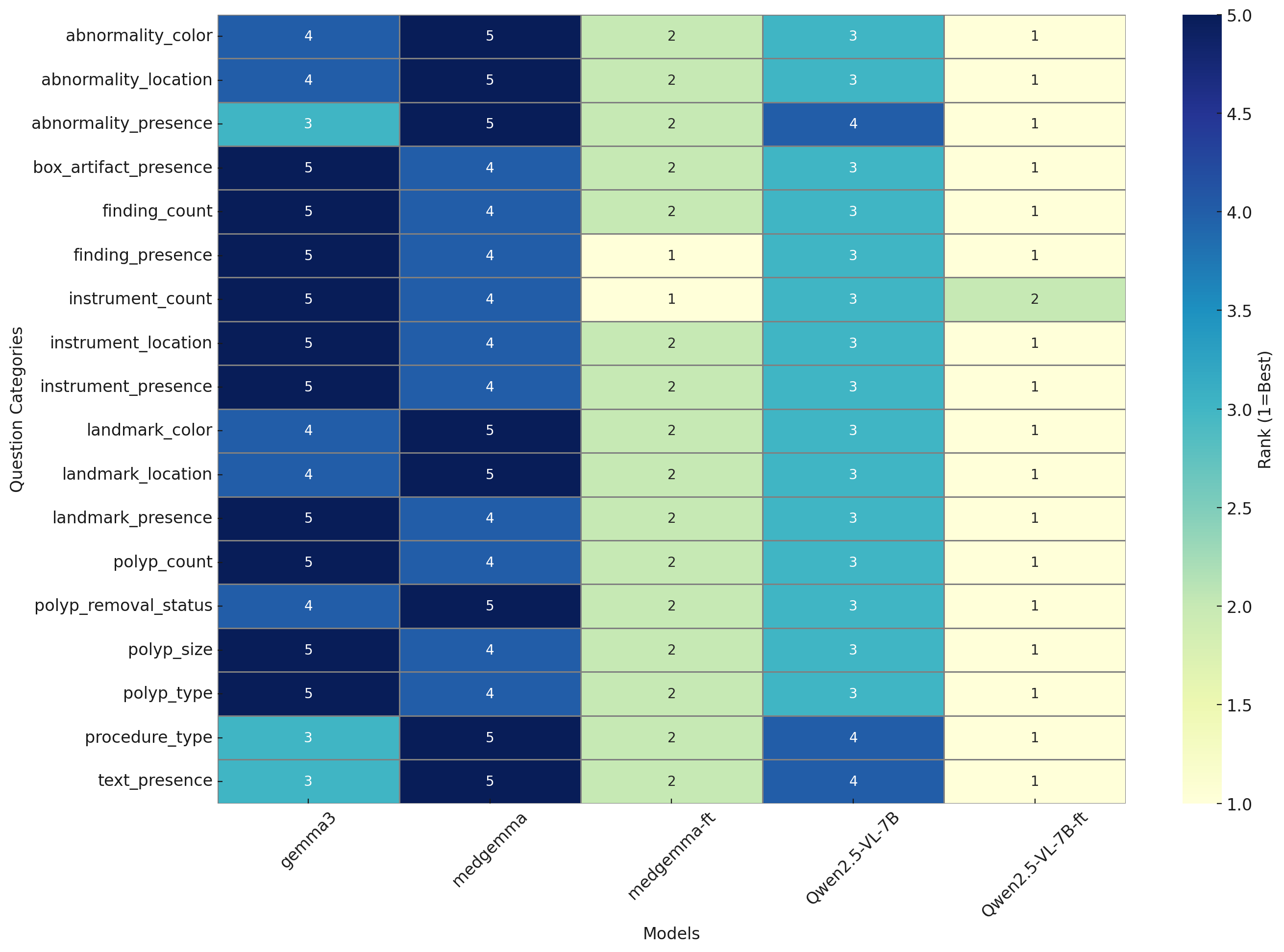
Figure 1: Rank-normalized heatmap illustrating comparative performance rankings (1 = best, 5 = worst) of the models across Kvasir-VQA categories. Qwen2.5-VL-7B-FT consistently ranks first across most categories.
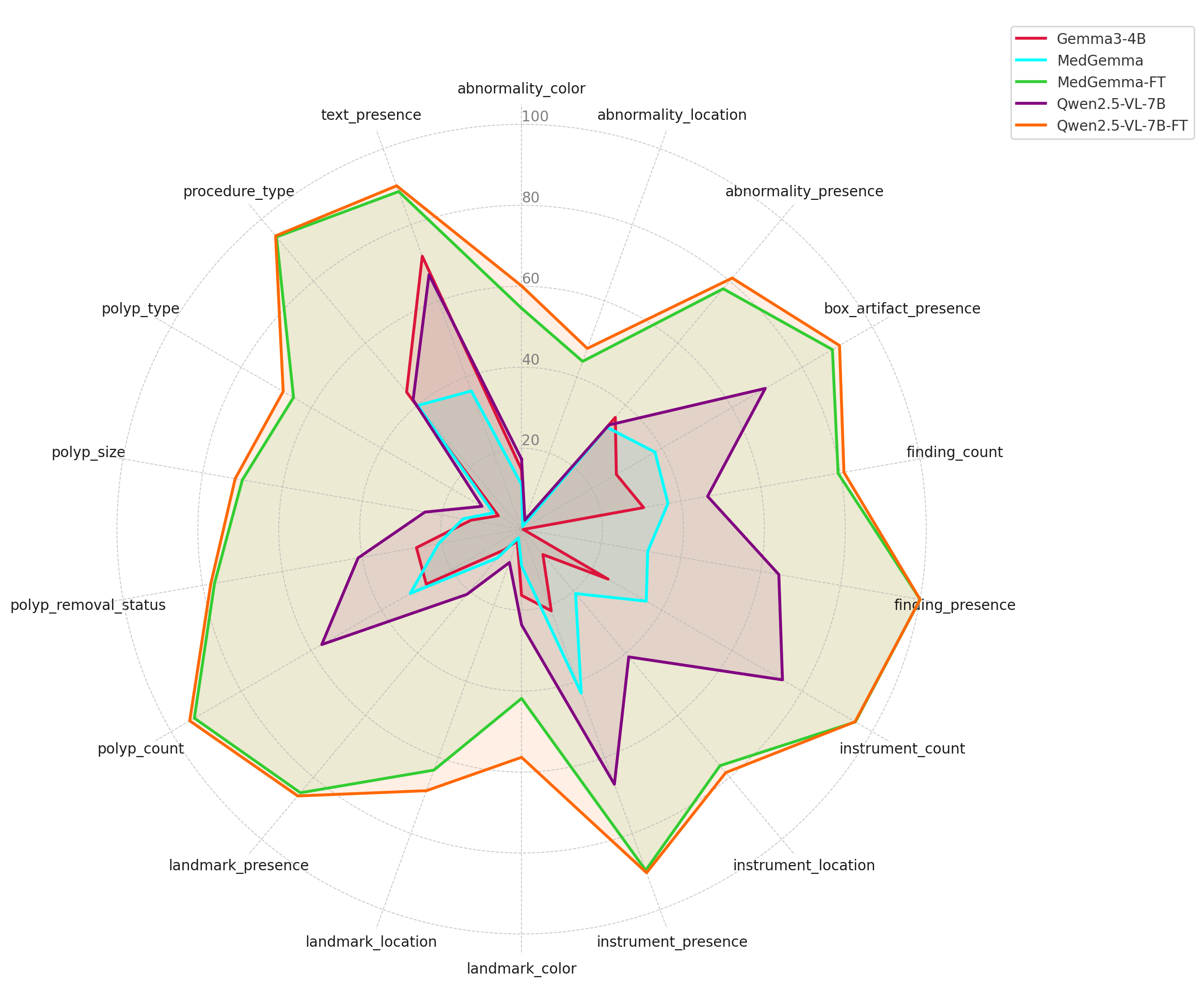
Figure 2: Radar plot showing absolute performance scores of five models (Gemma3-4B, MedGemma, MedGemma-FT, Qwen2.5-VL-7B, and Qwen2.5-VL-7B-FT) across various question categories. Higher values indicate better performance.
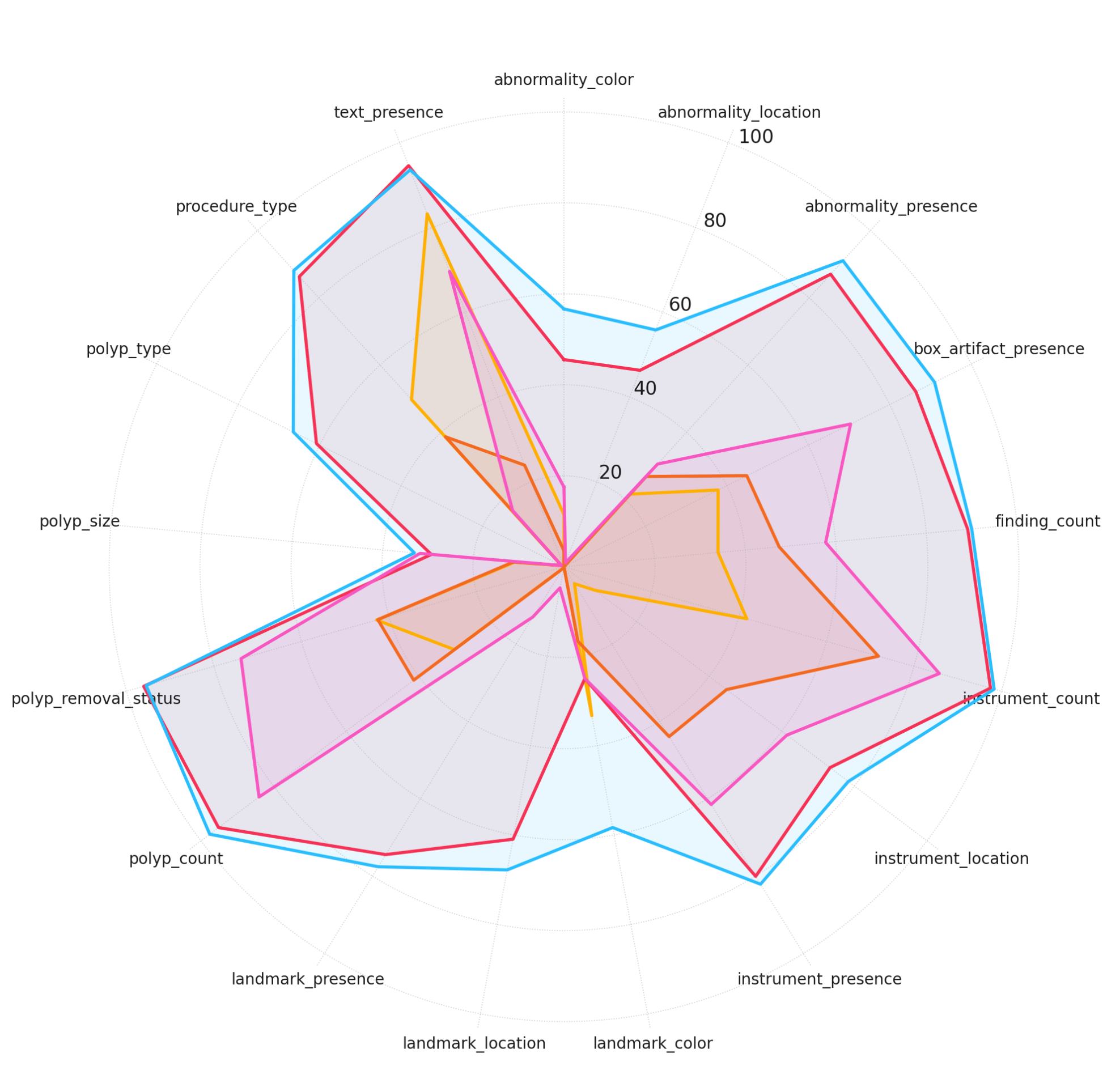
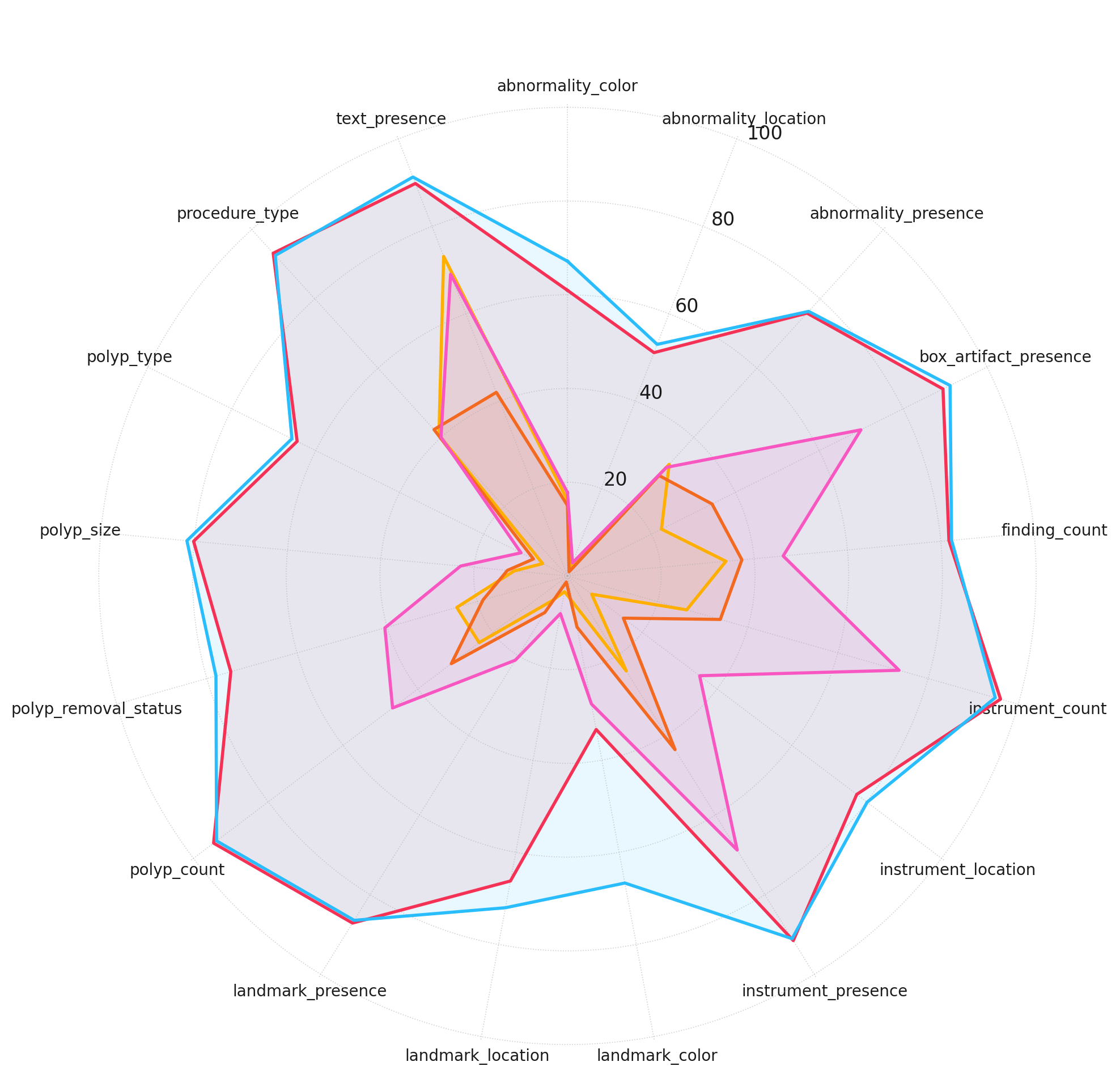
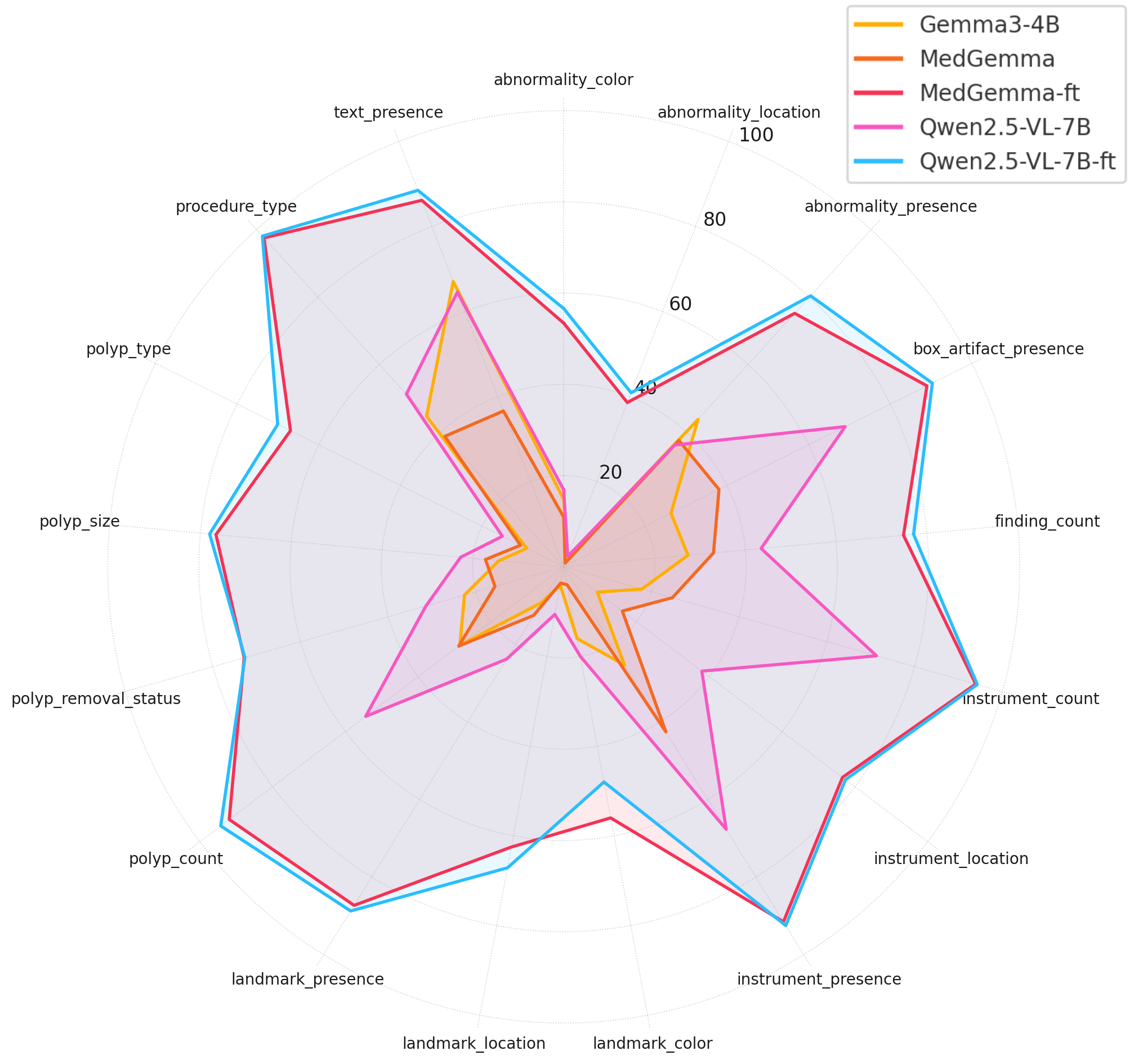
Figure 3: Model performance across different complexity levels. Accuracy scores are plotted for each model across different question categories, grouped by reasoning complexity.




