- The paper introduces MedNeXt-v2, a novel compound-scaled 3D ConvNeXt architecture that integrates GRN modules for stabilizing training in medical segmentation tasks.
- It employs depth, width, and context scaling, achieving up to +1.0 DSC improvement over state-of-the-art baselines across CT and MR datasets.
- The work emphasizes rigorous backbone selection and systematic benchmarking, enabling efficient representation learning and reliable transfer to clinical segmentation tasks.
MedNeXt-v2: Advancing Scalable Supervised Representation Learning for 3D Medical Image Segmentation
Introduction
The domain of 3D medical image segmentation has witnessed rapid shifts toward large-scale supervised representation learning, paralleling developments in general computer vision. However, many efforts remain constrained by legacy backbones and limited architectural benchmarking, with a prevailing focus on expanding dataset size. The paper "MedNeXt-v2: Scaling 3D ConvNeXts for Large-Scale Supervised Representation Learning in Medical Image Segmentation" (2512.17774) addresses systemic limitations by introducing MedNeXt-v2—a compound-scaled 3D ConvNeXt architecture designed for state-of-the-art representation learning. This work performs rigorous backbone selection prior to pretraining, integrates micro-architectural advances such as 3D Global Response Normalization (GRN), and systematically benchmarks MedNeXt-v2 against prominent public pretrained networks across diverse segmentation challenges.

Figure 1: MedNeXt-v2 sets a new state-of-the-art in 3D medical image segmentation, outperforming leading baselines across multiple 3D segmentation tasks.
Architectural Innovations and Scaling Strategies
MedNeXt-v2 is built upon the ConvNeXt foundation but compounds scaling across depth, width, and input context. The architecture incorporates 3D GRN modules to mitigate activation collapse prevalent in deep ConvNets, thereby fostering robust feature diversity throughout volumetric representation learning.
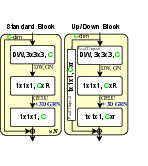
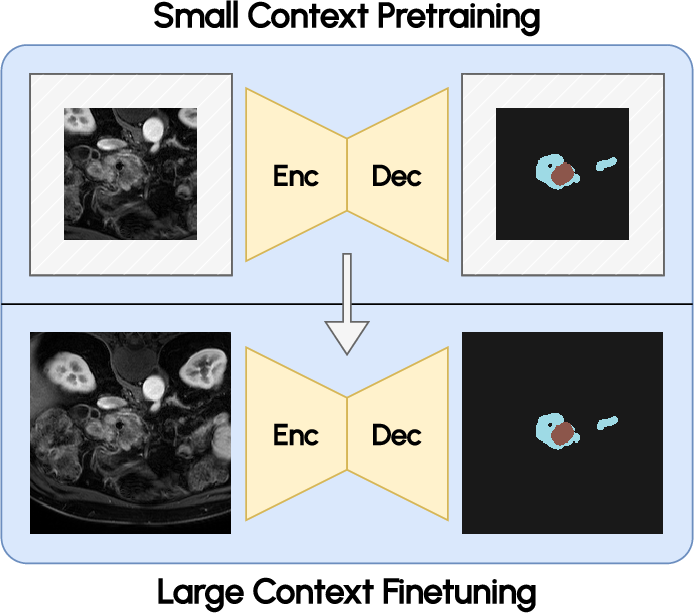
Figure 2: Micro-architecture of MedNeXt-v2, highlighting the integration of GRN and channel scaling strategies.
Noteworthy compounds of scaling in MedNeXt-v2 include:
- Depth scaling: Adoption of a deep 52-layer configuration, paralleling efficient scaling paradigms akin to EfficientNet.
- Width scaling: Doubling channel capacity at each network stage to enhance representational power.
- Context scaling: Fine-tuning with enlarged 1923 input patches amplifies spatial context, leading to notable downstream accuracy gains despite modest pretraining resource demands.
GRN is shown to consistently stabilize training dynamics and prevent dead or saturated channel activations. Channel activation visualization further evidences the role of GRN in sustaining informative, non-redundant features across varied anatomical and pathological targets.








Figure 3: Channel activation visualization—3D GRN in MedNeXt-v2 reduces redundant activations, preventing feature collapse.
Empirical Evaluation and Benchmarking
MedNeXt-v2 undergoes benchmarking on six challenging public datasets, totaling 144 structures spanning CT and MR modalities. Pretraining is conducted on 18,000 CT volumes covering 44 target structures, paired with rigorous fine-tuning regimens. The design enforces backbone validation against state-of-the-art baselines prior to scaling, ensuring optimal representation capabilities.
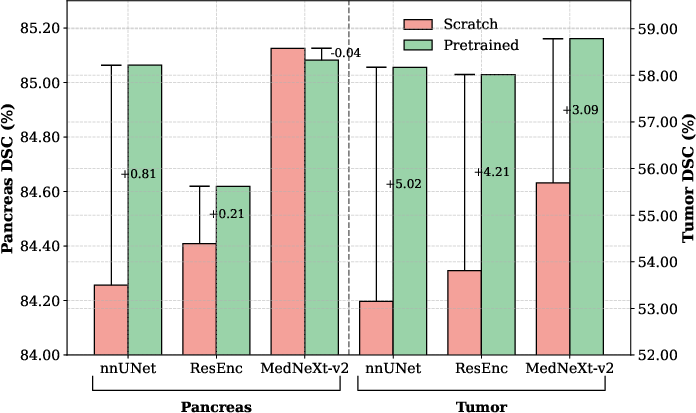
The incorporation of GRN distinctly improves performance both when training from scratch and after large-scale supervised pretraining. MedNeXt-v2 outperforms MedNeXt-v1 and alternative pretrained baselines—such as nnUNet, ResEncL, Vista3D, TotalSegmentator, MRSegmentator, and STU-Net—by up to +1.0 DSC on difficult datasets, including pediatric CT and pancreatic tumor segmentation.

Figure 4: MedNeXt-v2 outperforms MedNeXt-v1 from scratch and during fine-tuning, attributed to GRN stabilization.
Further, context scaling during fine-tuning yields substantial benefit, with larger patches enabling superior segmentation at anatomical boundaries and improving performance metrics relative to both smaller patch variants and competing public models.
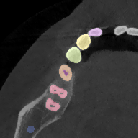


Figure 5: Increased context during fine-tuning with 1923 patches enables more accurate segmentation near image boundaries.
Qualitative analysis demonstrates MedNeXt-v2's capability in both CT and MR domains, with notable improvements in boundary delineation and fine anatomical detail.
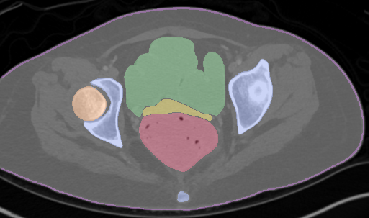
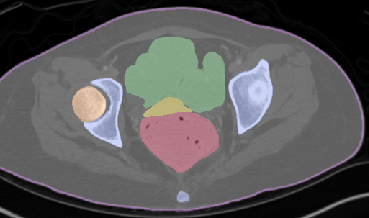




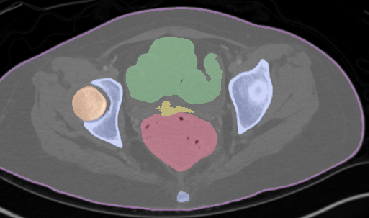




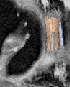




Figure 6: Qualitative visualizations of MedNeXt-v2 and context-scaled variant on CT and MR datasets, compared against public baselines.
Insights from Systematic Benchmarking
Comprehensive benchmarking leads to several impactful findings:
MedNeXt-v2 Macro Architecture
MedNeXt-v2 features a symmetric 5-hierarchy UNet-style architecture with compound scaling and residual ConvNeXt blocks at each stage. GRN is integrated throughout all residual paths and up/downsampling layers for effective learning dynamics.

Figure 8: MedNeXt-v2 macro-architecture parallels MedNeXt-v1, but augments representation via GRN and scalable design.
Implications and Future Directions
MedNeXt-v2 conclusively demonstrates that strategic backbone selection, combined with sensible micro-architectural innovation, is pivotal for the advancement of large-scale 3D medical image segmentation models. The findings prompt a methodological shift from dataset size-centric practices toward representation-aware architecture benchmarking and scaling. Practical implications include more reliable transfer learning for downstream clinical segmentation tasks and rapid fine-tuning pipelines with minimized resource footprint.
Theoretically, the work substantiates the role of architectural bias—such as ConvNeXt-style inductive properties—over mere parameter count or data scale, particularly in data-constrained and annotation-sparse regimes characteristic of medical imaging. Future research may explore adaptive scaling strategies in response to domain-specific saturation, further cross-modality generalization, or integration with semi/self-supervised pipelines as annotation burdens persist.
Conclusion
MedNeXt-v2 sets a new standard in 3D supervised medical image segmentation, combining rigorous backbone validation, micro-architectural refinement via GRN, and efficient scaling strategies. Its systematic benchmarking against state-of-the-art pretrained models reaffirms the importance of representation-centric design. The work lays a robust foundation for optimized segmentation pipelines and motivates continued research in scalable, adaptive architectures for medical AI.