- The paper introduces Dino U-Net, integrating a frozen DINOv3 encoder with a U-Net decoder via a fidelity-aware projection module for high-precision segmentation.
- It demonstrates superior performance, achieving a +1.87 Dice improvement and -3.04 HD95 reduction across seven diverse medical imaging datasets.
- By freezing the encoder and training only lightweight modules, the approach ensures parameter efficiency and robust generalization in 2D medical image segmentation.
Dino U-Net: Leveraging High-Fidelity Dense Features from Foundation Models for Medical Image Segmentation
Introduction
Dino U-Net introduces a hybrid encoder-decoder architecture that integrates a frozen DINOv3 vision transformer as the encoder with a U-Net decoder, targeting high-precision medical image segmentation. The central innovation lies in the exploitation of high-fidelity, dense features from DINOv3, a self-supervised vision foundation model, and the design of a fidelity-aware projection module (FAPM) to preserve and adapt these features for downstream segmentation. The architecture is evaluated across seven diverse medical imaging datasets, demonstrating consistent improvements over state-of-the-art baselines in both segmentation accuracy and parameter efficiency.
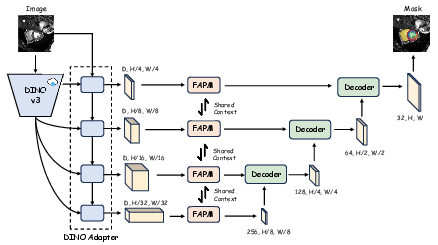
Figure 1: Overview of the Dino U-Net architecture, highlighting the frozen DINOv3 encoder, adapter, FAPM, and U-Net decoder. Only the adapter, FAPM, and decoder are trainable.
Architecture and Methodology
Encoder: Frozen DINOv3 with Adapter
The encoder leverages a frozen DINOv3 backbone, pre-trained on large-scale natural image data. To bridge the domain gap and fuse semantic and spatial information, an adapter module is introduced. The adapter employs a dual-branch design: a spatial prior module (SPM) extracts low-level spatial features, while the DINOv3 backbone provides high-level semantic features. These are fused via deformable cross-attention in a series of interaction blocks, producing multi-scale, semantically enriched feature maps.
Fidelity-Aware Projection Module (FAPM)
The FAPM addresses the challenge of dimensionality reduction from the high-dimensional DINOv3 features to the lower-dimensional space required by the U-Net decoder. It operates in two stages:
- Orthogonal Decomposition: Multi-scale features are projected into a shared low-rank space using a 1x1 convolution, separating scale-invariant context from scale-specific details.
- Progressive Refinement: Each scale undergoes further refinement via depthwise separable convolutions, squeeze-and-excitation (SE) blocks, and residual connections, restoring expressive capacity and ensuring high-fidelity feature transfer.
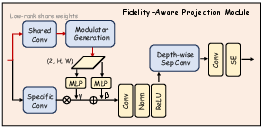
Figure 2: Detailed architecture of the FAPM, illustrating the orthogonal decomposition and progressive refinement stages.
Decoder: Standard U-Net
The decoder is a conventional U-Net, receiving the refined, multi-scale skip connections from the FAPM. This design enables precise localization and boundary delineation, leveraging the rich representations from the encoder.
Experimental Evaluation
Datasets and Baselines
Dino U-Net is evaluated on seven public datasets spanning endoscopy, fundus imaging, ultrasound, microscopy, cardiac MRI, prostate MRI, and surgical endoscopy. Baselines include nnU-Net, SegResNet, UNet++, U-Mamba, U-KAN, Swin U-Mamba, and SAM2-UNet, covering both conventional and foundation model-based approaches.
Implementation Details
All experiments are conducted in PyTorch, following nnU-Net's standardized preprocessing, data augmentation, and training protocols. Only the adapter, FAPM, and decoder are trainable; the DINOv3 backbone remains frozen. The models are trained with a combined Dice and cross-entropy loss, using Adam optimizer and polynomial learning rate decay. Inference employs a sliding window with Gaussian weighting.
Quantitative and Qualitative Results
Dino U-Net consistently outperforms all baselines across datasets in both Dice Similarity Coefficient and 95% Hausdorff Distance (HD95). The 7B-parameter variant achieves the highest average Dice (76.43%) and lowest average HD95 (18.76), with improvements of +1.87 Dice and -3.04 HD95 over the best baseline. Notably, even the smallest Dino U-Net S model (5.1M parameters) surpasses all baselines in average performance, demonstrating strong parameter efficiency.
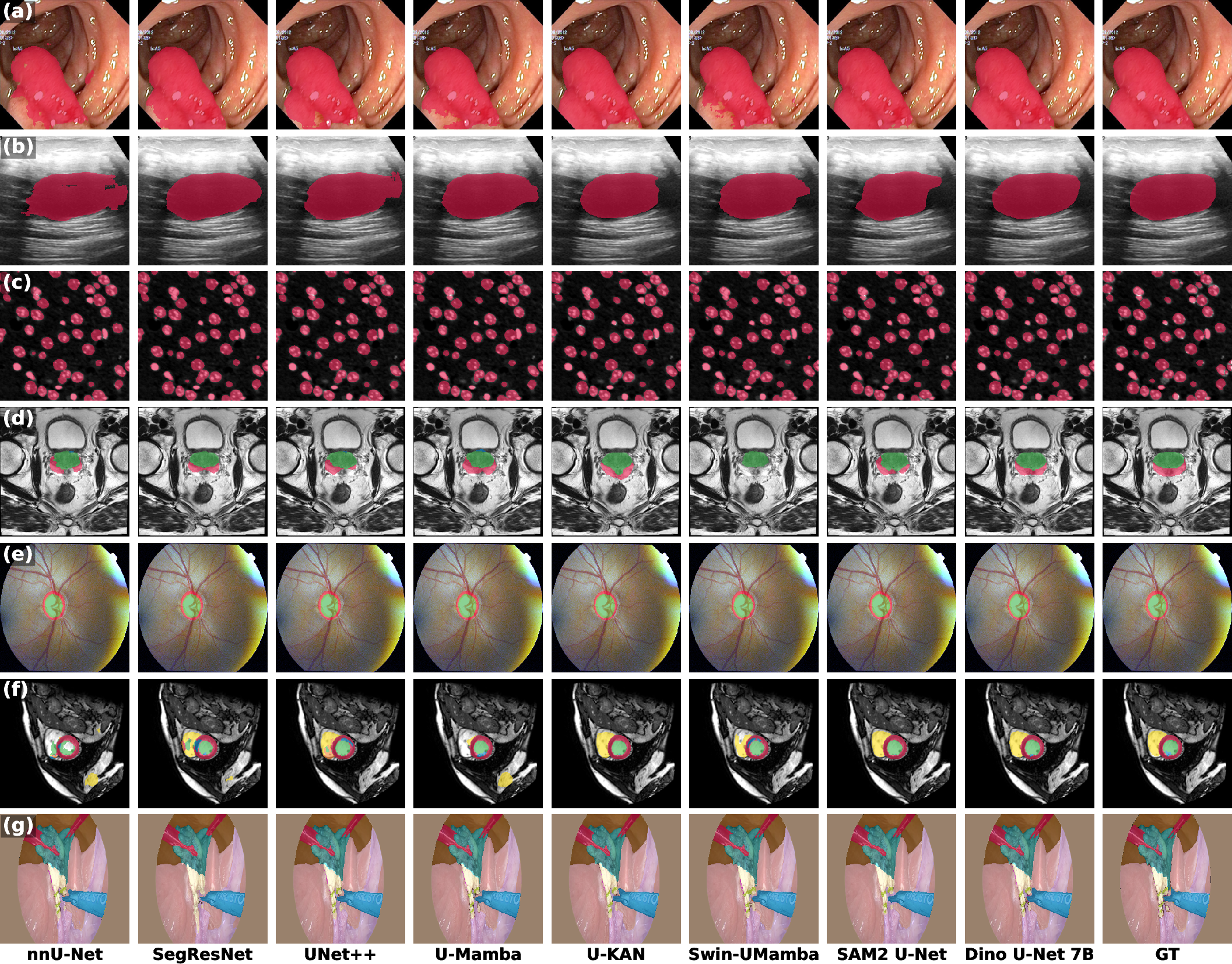
Figure 3: Qualitative comparison of segmentation results across seven datasets, showing Dino U-Net's superior boundary delineation and completeness.
Ablation Study: FAPM
Ablation experiments confirm the critical role of FAPM. Removing FAPM and replacing it with simple 1x1 convolutions leads to a 0.56–0.79% drop in Dice and a 0.09–1.75mm increase in HD95, particularly impacting boundary precision. For the largest model, FAPM also reduces parameter count due to its parameter-sharing mechanism.
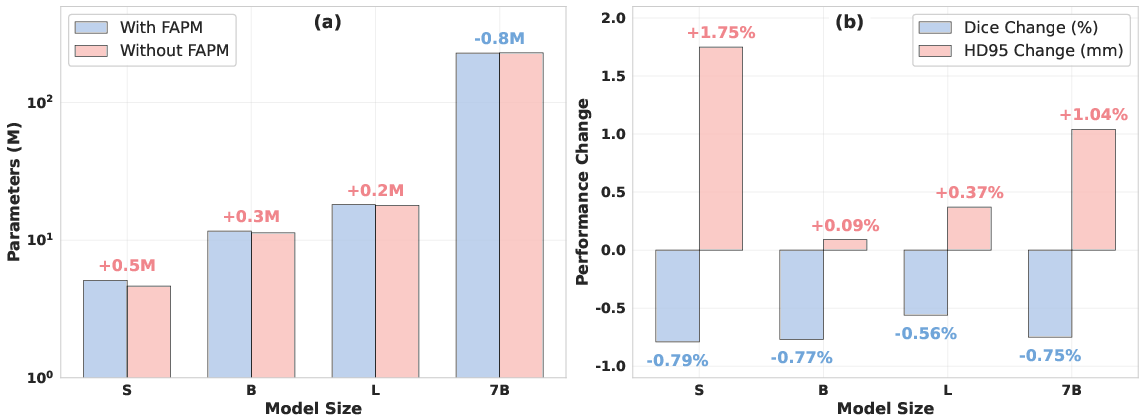
Figure 4: Ablation paper results, demonstrating FAPM's impact on parameter efficiency and segmentation accuracy.
Discussion
The results substantiate several key claims:
- General-Purpose Features vs. Segmentation-Specific Features: DINOv3's self-supervised, general-purpose representations provide a more robust inductive bias for medical segmentation than segmentation-optimized encoders like SAM. This is attributed to DINOv3's ability to capture texture, context, and material properties, which are critical for differentiating ambiguous or low-contrast anatomical structures.
- Scalability: Segmentation performance scales with the size of the DINOv3 backbone, with no observed saturation up to 7B parameters. This suggests that further scaling of foundation models may yield additional gains in medical imaging tasks.
- Parameter Efficiency: By freezing the encoder and training only lightweight modules, Dino U-Net achieves state-of-the-art results with fewer trainable parameters, reducing overfitting risk and computational burden during training.
- FAPM's Role: The fidelity-aware projection is essential for preserving high-fidelity features during dimensionality reduction, directly impacting boundary accuracy and overall segmentation quality.
Limitations and Future Directions
- 2D Limitation: The current architecture is restricted to 2D segmentation. Extending to 3D volumetric data (e.g., CT, MRI) is a logical next step.
- Inference Cost: While training is efficient, inference with the 7B backbone is resource-intensive. Model distillation or pruning could mitigate this.
- Generalization: Further evaluation on rare pathologies and cross-domain transfer would strengthen claims of generalization.
Conclusion
Dino U-Net demonstrates that integrating a frozen, large-scale self-supervised vision foundation model with a U-Net decoder, augmented by a fidelity-aware projection module, yields superior and efficient medical image segmentation. The approach leverages the representational power of billion-parameter models while maintaining practical training requirements. This work suggests that future advances in medical imaging will increasingly depend on the effective adaptation and projection of foundation model features, with further gains likely as model scale and projection techniques evolve.



