Entangled Schrödinger Bridge Matching (2511.07406v1)
Abstract: Simulating trajectories of multi-particle systems on complex energy landscapes is a central task in molecular dynamics (MD) and drug discovery, but remains challenging at scale due to computationally expensive and long simulations. Previous approaches leverage techniques such as flow or Schrödinger bridge matching to implicitly learn joint trajectories through data snapshots. However, many systems, including biomolecular systems and heterogeneous cell populations, undergo dynamic interactions that evolve over their trajectory and cannot be captured through static snapshots. To close this gap, we introduce Entangled Schrödinger Bridge Matching (EntangledSBM), a framework that learns the first- and second-order stochastic dynamics of interacting, multi-particle systems where the direction and magnitude of each particle's path depend dynamically on the paths of the other particles. We define the Entangled Schrödinger Bridge (EntangledSB) problem as solving a coupled system of bias forces that entangle particle velocities. We show that our framework accurately simulates heterogeneous cell populations under perturbations and rare transitions in high-dimensional biomolecular systems.
Paper Prompts
Sign up for free to create and run prompts on this paper using GPT-5.
Top Community Prompts
Explain it Like I'm 14
What is this paper about?
This paper introduces a new way to simulate how many moving things (like atoms in a protein or groups of cells) change over time when they influence each other. The authors call their method Entangled Schrödinger Bridge Matching (EntangledSBM). It helps “steer” these systems from a starting state to a desired target state, even when the parts interact in complex, changing ways.
What questions does the paper try to answer?
The paper focuses on three main questions:
- How can we predict and simulate the paths of many interacting particles (or cells) when each one’s movement depends on the others?
- How can we do this efficiently using only snapshots (static data points), instead of running super long and expensive simulations?
- Can the method learn to reach both known target states and new, unseen target states without retraining?
How did the authors approach the problem?
Think of a group of people trying to walk through a crowded city from point A to point B. Each person’s speed and direction change based on the people around them (to avoid bumping into others), and the environment (like roads and obstacles). The authors build a “smart guidance system” that adds gentle steering to each person so the whole group ends up at the target location in a realistic, efficient way. Here’s how they do it:
Modeling moving particles: base dynamics
- The system has many particles (like atoms or cells), each with a position (where it is) and a velocity (how fast and in what direction it’s moving).
- The natural motion is described by rules known as “Langevin dynamics.” These rules model:
- Forces that come from the environment (like energy hills and valleys).
- Random small kicks (like bumps from surrounding molecules).
- Without steering, particles follow these rules and wander around according to the energy landscape.
The Schrödinger bridge idea: best paths from start to target
- Imagine you want the group to end at a target area (a desired distribution of states) without dictating an exact path.
- A Schrödinger bridge finds the most likely “bridge” of paths from the start distribution to the target distribution, balancing realism and guidance.
- The method learns a “bias force” (a steering force) that gently tilts the natural motion toward the target.
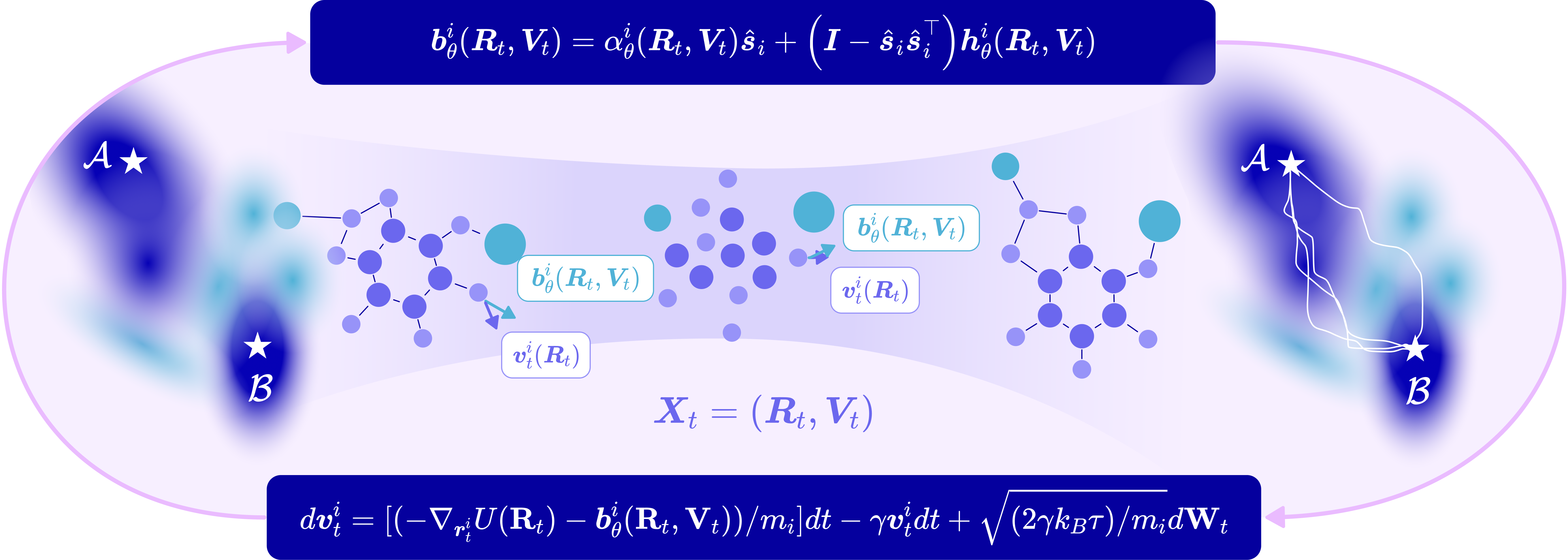
Entangled bias force: making interactions matter
- In many systems (like proteins or heterogeneous cell populations), parts don’t just act independently. One particle’s movement depends on others and changes over time.
- The authors introduce an “entangled bias force” that depends on:
- The positions of all particles.
- The velocities of all particles.
- This means every particle’s steering is aware of what the others are doing, so the system behaves more realistically.
Learning with Transformers: seeing the whole group
- They use a Transformer (a neural network architecture originally used for language) to let each particle “attend” to all others.
- Each particle is like a token with features (its position and velocity), and the Transformer mixes information across all particles.
- For molecules, they align coordinates using a standard method (Kabsch) so the model is not confused by overall rotations or shifts.
A safe steering rule: don’t move away from the target
- To avoid steering in the wrong direction, they constrain part of the bias force to always have a non-negative projection toward the target.
- Intuitively: the steering always includes a “forward” component toward the goal, while allowing “sideways” adjustments to avoid obstacles or make group movements smoother.
Training objective: learn from good paths, off-policy
- The method uses ideas from optimal control (planning the best way from A to B with minimal extra effort).
- They introduce a cross-entropy objective (a way to score paths) that is stable and has a unique best solution.
- Practically, they:
- Simulate batches of paths with the current steering.
- Score how well each path ends up near the target (give it a “reward”).
- Weight the better paths more (importance weighting), store them in a replay buffer, and reuse them to improve the steering.
- This “off-policy” approach means they don’t need to throw away past simulations—they learn efficiently from what they already ran.
How did they test it?
They tested EntangledSBM on two challenging tasks:
- Cell clusters under drug perturbations: predicting how groups of cells change state when a drug is applied.
- Protein transition paths: simulating rare, realistic motion that carries a protein from one shape to another over a complicated energy landscape.
What did they find?
Cells under drug perturbations
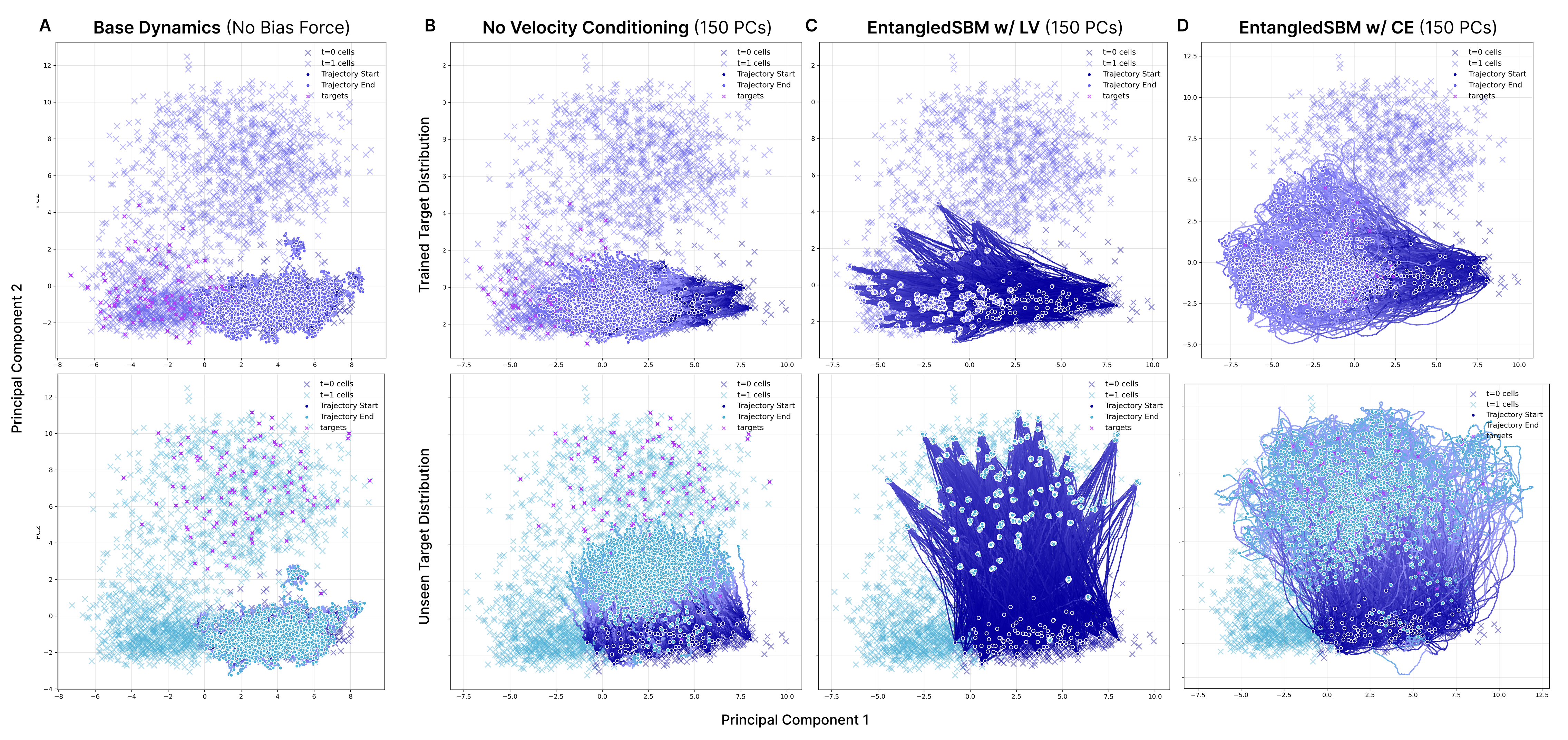
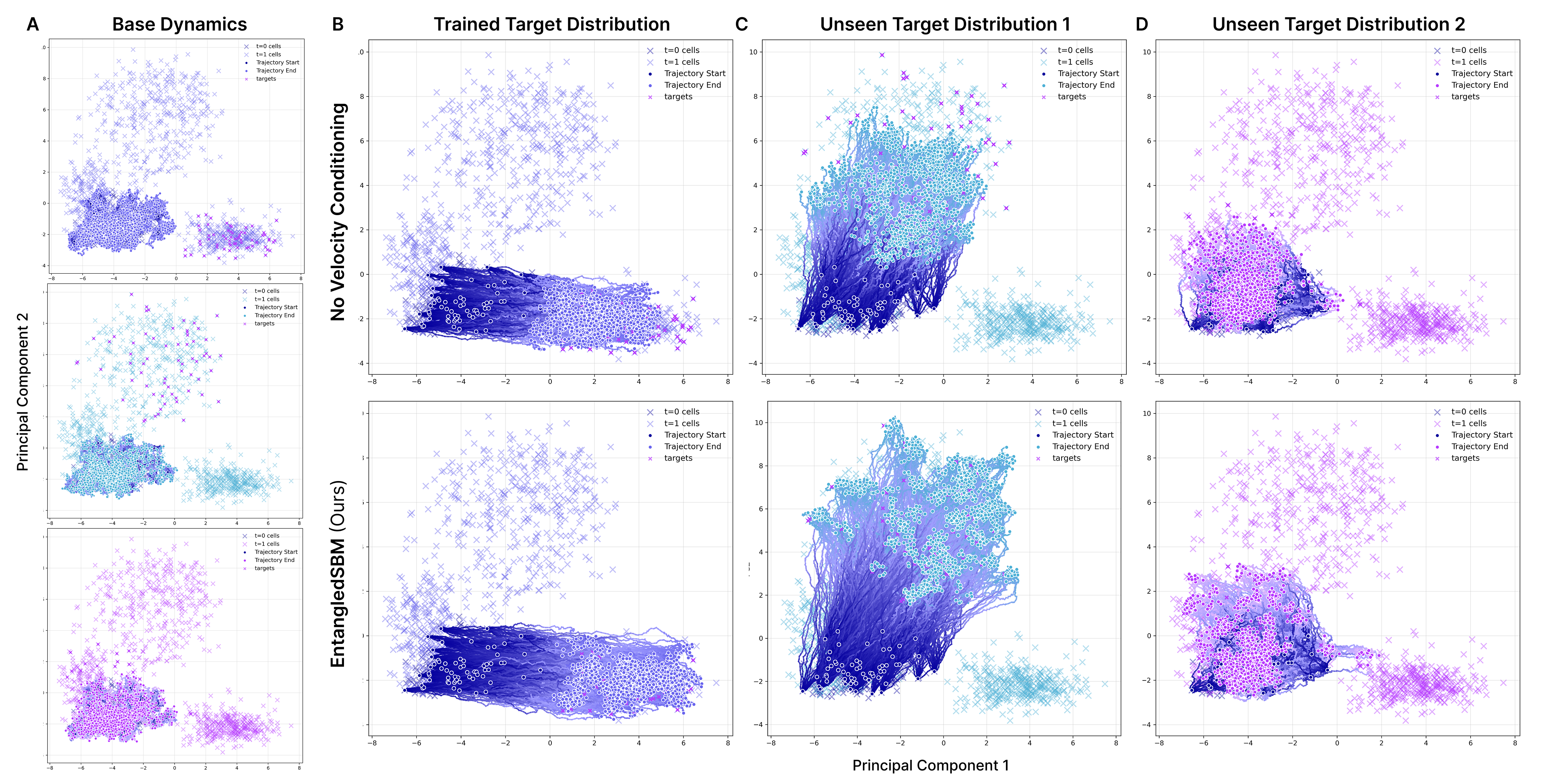
- Their method accurately guided clusters of cells from healthy (unperturbed) states to drug-affected (perturbed) states.
- It reconstructed the target distributions (how the cells should look) across many dimensions (principal components, or PCs).
- Importantly, it generalized to unseen target cell populations (perturbation clusters not used during training), showing it can adapt to new targets.
- Conditioning the steering on both positions and velocities was crucial—models that ignored velocities performed worse.
- Their cross-entropy training produced smooth, realistic paths along the data manifold (not abrupt jumps), matching how cells transition biologically.
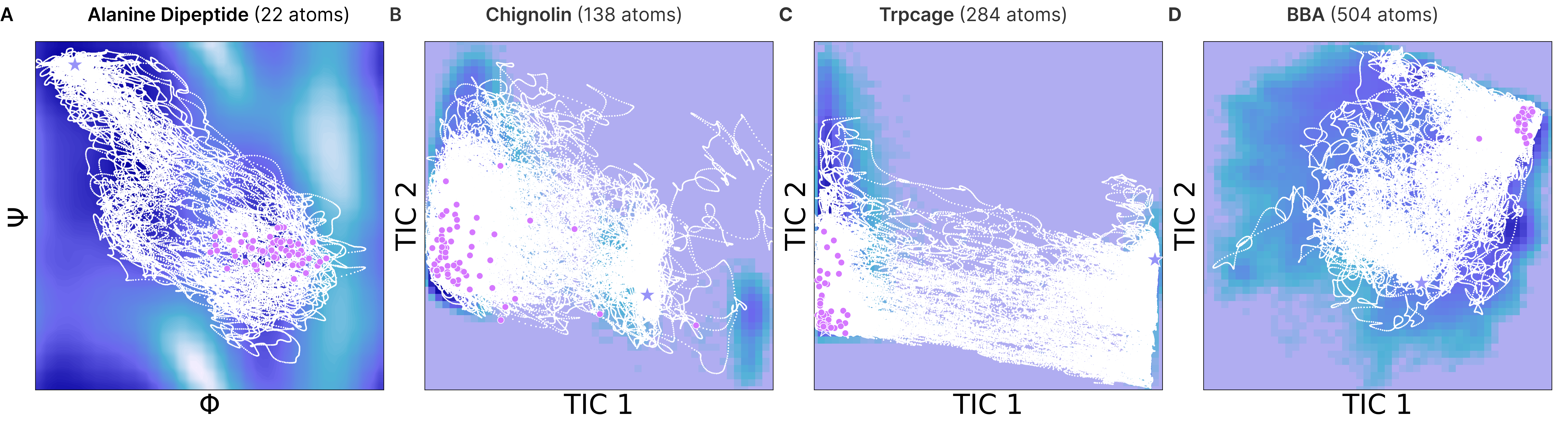
Protein transition paths (molecular dynamics)
- They simulated high-resolution, feasible transition paths for several systems (like Alanine Dipeptide and fast-folding proteins).
- Their method achieved:
- Higher target hit rates (more paths successfully reaching the target state).
- Lower RMSD (closer final structures to the target) compared to multiple baselines.
- Compared with traditional methods (like steered MD), EntangledSBM often found better routes through complex energy landscapes, even with many atoms, where rare transitions are hard to simulate.
Why is this important?
- Many real-world systems are made of parts that influence each other in complicated, changing ways: molecules in a cell, cells in a tissue, or even tokens in a sequence.
- Standard methods often assume every part behaves like the average (mean-field), which breaks down when the system is diverse or strongly interactive.
- EntangledSBM captures these interactions directly by letting velocities and positions across the whole group shape the steering.
- It works from snapshots (not needing huge, long simulations), can generalize to new targets, and produces smooth, realistic paths.
What could this impact?
- Drug discovery: predict how different cell populations respond to new drugs, even with limited data.
- Molecular design: explore feasible protein transitions and rare events that are hard to reach with brute-force simulations.
- Complex systems modeling: any setting with many interacting agents (biological, physical, or even computational), where realistic, controllable trajectories matter.
In short, EntangledSBM is a smarter way to “guide crowds” of interacting particles from where they are to where we want them to go—safely, efficiently, and in a way that respects how they influence one another.
Knowledge Gaps
Knowledge gaps, limitations, and open questions
Below is a consolidated list of concrete gaps and unresolved questions that the paper leaves open. These points can guide follow-up theoretical, methodological, and empirical work.
Theory and objectives
- Availability and estimation of the score of the target distribution: the bias-force parameterization requires the per-particle target score ∇ log π_B, but for indicator-type targets (e.g., relaxed sets around a state) or empirical target samples (e.g., cell clusters), this score is undefined or unavailable. The paper does not specify how to estimate it (e.g., score model, KDE, energy-based model), nor analyze the impact of score approximation errors on guarantees.
- Non-increasing distance guarantee applicability: Proposition relies on small Δt and a per-particle direction toward a concrete target state R_B, yet many experiments use distributional targets. It remains unclear how R_B is chosen within π_B, whether the guarantee holds uniformly across a distribution, and how it extends when π_B is multi-modal.
- Existence/uniqueness and well-posedness of the EntangledSB problem: conditions (e.g., Lipschitz, linear growth) ensuring existence and uniqueness of solutions for the entangled, second-order controlled SDEs are not stated. The validity of Girsanov’s change-of-measure under entangled drifts depending on all positions and velocities is not established.
- From convexity in path measures to convergence in parameters: while the cross-entropy (CE) objective is convex in the path measure, this does not imply convexity in network parameters θ. No convergence guarantees are provided for function approximation with off-policy, self-normalized importance weighting and replay buffers.
- Control optimality vs. path realism trade-offs: the paper does not analyze how the quadratic control cost and terminal reward shape path realism (e.g., avoiding high-energy regions) nor provide conditions ensuring low-energy, physically plausible bridges under the entangled bias.
- Terminal constraints in second-order dynamics: targets are imposed on positions only, while second-order dynamics evolve in phase space. The implications of leaving terminal velocity unconstrained (e.g., mismatch between desired phase-space bridge and position-only constraints) are not analyzed.
- Impact of discretization and stochastic calculus choices: the discrete-time objective reconstructs ΔW_k from stored steps, but discretization bias (Itô vs. Stratonovich, strong/weak order) and its effect on the learned control and guarantees are not studied.
Method and modeling choices
- Estimation of “unconditional velocities” for snapshot data: the framework uses unconditional velocities V_t(R_t) in training/simulation, but the paper does not specify how these are obtained from static datasets (e.g., RNA velocity models, learned drifts, finite differences on a manifold), nor quantify their noise sensitivity.
- Constructing π_B for cell populations: the target distribution for perturbed cells is used in training, but the method to represent π_B (density model, score estimator, kernel density, classifier-free guidance) is not described, making the implementation of the score-based parameterization unclear.
- Orthogonal component unconstrained: the orthogonal bias component can induce large sideways motion. There is no constraint set to preserve physical invariants (e.g., bond lengths, angular momentum, rigid constraints), potentially yielding unphysical MD trajectories.
- Symmetry handling in MD: the approach uses Kabsch alignment to a target for frame invariance but does not adopt an E(3)-equivariant architecture. The impact on generalization across orientations/conformations and on enforcing physical symmetries remains unexplored.
- Particle exchangeability and identity: the transformer does not explicitly enforce permutation invariance (relevant for exchangeable particles such as cells). The implications for heterogeneous vs. homogeneous particle systems are not analyzed.
- Sensitivity to control hyperparameters: the method depends on Δt, temperature schedules (annealing), friction γ, and control scaling (softplus α). There is no sensitivity or stability analysis to guide practitioners.
- Importance-weight approximation and stability: the paper replaces the intractable normalization Z with a batch softmax approximation; the bias/variance, stability under heavy-tailed rewards, and replay-buffer staleness are not characterized. No variance reduction or effective sample size diagnostics are reported.
- Replay buffer and off-policy reuse: how to prevent bias from stale trajectories, weight collapse, or divergence due to policy lag is not addressed (e.g., target networks, trust regions, partial on-policy refresh).
- Entanglement strength and architecture ablations: the contribution of velocity conditioning vs. position-only, the benefit of attention-based entanglement vs. mean-field or graph-based alternatives, and the value of the orthogonal component are not systematically ablated.
- Conditioning on target descriptions: generalization relies on feeding direction and distance to target; for distributional targets, how to parameterize conditioning (e.g., summary statistics, learned embeddings) and how far OOD conditioning remains reliable are open.
Experiments and evaluation
- Physical plausibility of MD paths: for some proteins, the reported highest-energy transition state (ETS) is much higher than baselines, raising concerns about physical realism (e.g., barriers surmounted via nonphysical work). No checks such as committor analysis, reactive flux, work distributions, or constraint violations are reported.
- Scaling to larger systems: results are limited to small proteins and cell clusters of n=16. There is no analysis of computational complexity (O(n2) attention), memory requirements for long trajectories, or scaling behavior to hundreds/thousands of particles.
- Limited baselines for interacting dynamics: comparisons focus on SMD/PIPS/TPS-DPS for MD and a position-only variant for cells. There is no comparison to mean-field interaction models, graph neural simulators, or E(3)-equivariant control policies capturing interactions.
- Biological plausibility and mechanistic validation: cell experiments evaluate end-point distribution matching (RBF-MMD, Wasserstein) but do not assess intermediate biological realism (e.g., lineage consistency, gene program trajectories, known perturbation-response markers).
- Generalization beyond closely related targets: the “unseen” targets are clustered from the same perturbation dataset. There is no stress test for targets that are far from training distribution, across different perturbation types or cell lines, nor calibration of failure modes.
- Data efficiency and label requirements: the method’s dependence on the amount and quality of perturbed data per target (and how it scales when targets are scarce) is not quantified.
- Uncertainty and multi-modality of paths: the framework does not quantify uncertainty over path ensembles or support sampling multiple distinct modes in a controlled way (e.g., via conditional mixture policies or posterior predictive checks).
- Long-horizon credit assignment: no paper of how performance degrades with horizon length, nor techniques to mitigate vanishing/exploding gradients in path-space learning.
- Runtime and resource reporting: wall-clock cost, simulation-to-optimization ratio, and throughput vs. baselines are not reported, limiting practical adoption.
- Robustness to noisy/mis-specified targets: the impact of target misspecification, noisy labels, or adversarial perturbations in π_B on the learned control and generated paths is not evaluated.
- Branching and multi-target conditioning: although related work on branching SB is cited, the proposed method is not evaluated on branching/multi-terminal settings or mixtures of targets within a single run.
Extensions and integration opportunities
- Incorporating physics-informed constraints: adding hard/soft constraints (distance, angle, energy barriers), conservative fields, or symplectic structure could improve MD realism; the paper does not explore such integrations.
- Equivariant and graph-based architectures: evaluating E(3)-equivariant transformers/graph nets for MD and permutation-invariant message passing for cell populations could enhance inductive biases for interactions.
- Score modeling for targets: training a separate score model for π_B (e.g., diffusion/EBM) to enable a principled and scalable implementation of the “direction to target” and to reconcile the theory (∇ log π_B) with practice.
- Multi-objective training: adding intermediate-state rewards (e.g., manifold adherence, energy penalties, biological pathway consistency) could reduce shortcut paths; how to design and balance these terms remains open.
Practical Applications
Immediate Applications
The following applications can be deployed today using the paper’s methods and results, with reasonable engineering effort and domain integration.
- Transition path sampling (TPS) for biomolecules in research pipelines
- Description: Use EntangledSBM to generate feasible, high–target-hit transition paths between metastable states (e.g., folding/unfolding, ligand binding/unbinding, allosteric transitions), leveraging its target-conditioned bias force and off-policy cross-entropy training. Demonstrated superior THP/RMSD vs. baselines on multiple proteins.
- Sector(s): Healthcare/biotech; Software (MD simulation).
- Tools/Workflow: OpenMM plugin or wrapper (“EntangledSBM-TPS”) to sample path ensembles at set temperatures; optional post-processing with umbrella sampling/metadynamics for free-energy refinement; TICA/dihedral angle feature pipelines; Kabsch alignment support.
- Assumptions/Dependencies: Clear definitions of initial/target states or a tractable target distribution; valid Langevin dynamics regime; sufficient compute (GPU for Transformer, CPUs/GPUs for MD); stable importance weights (batch softmax approximation of Z); small step size to respect non-increasing-distance guarantee.
- In-silico prediction of drug perturbation effects on heterogeneous single-cell populations
- Description: Forecast cell-cluster trajectories from unperturbed to perturbed distributions; generalizes to unseen clusters by conditioning the entangled bias on target statistics. Shown effective on Tahoe-100M (Clonidine, Trametinib) with improved reconstruction metrics and smoother manifold-respecting paths.
- Sector(s): Healthcare (drug discovery, translational research); Academia (computational biology).
- Tools/Workflow: Integration with Scanpy/Seurat pipelines; PCA/HVG preprocessing; data-manifold energy estimation; batch simulation of nearest-neighbor clusters (n≈16) under bias; ranking drugs/conditions by alignment to desired perturbed distributions.
- Assumptions/Dependencies: Quality density estimation for the data manifold; availability of representative unperturbed and perturbed snapshots; target distribution gradient or direction features; careful handling of batch effects; compute for Transformer and replay-buffer training.
- Guided seeding for enhanced sampling in MD
- Description: Use EntangledSBM paths to initialize or bias windows for umbrella sampling/metadynamics, reducing burn-in and improving coverage of rare transition channels.
- Sector(s): Healthcare/biotech; Materials science.
- Tools/Workflow: Generate ensembles of transition hypotheses; select representative intermediates as windows; run standard enhanced sampling and reweighting for thermodynamic correctness.
- Assumptions/Dependencies: EntangledSBM paths are used as proposals (not final thermodynamic estimates); accurate collective variables (CVs) or structural descriptors; downstream reweighting remains necessary.
- Allosteric pathway and conformational-intermediate discovery
- Description: Identify collective motions and plausible intermediate states along entangled multi-atom velocities to hypothesize allosteric pathways and design experiments.
- Sector(s): Structural biology, drug discovery.
- Tools/Workflow: Path ensemble clustering; contact-map changes along trajectories; network analysis of collective motions; cross-validation with cryo-EM/NMR intermediates.
- Assumptions/Dependencies: Availability of reasonable target states (e.g., active/inactive conformations); mapping of CVs to biology; validation via orthogonal experiments.
- Data augmentation for ML models in structural biology and single-cell analysis
- Description: Generate physically plausible intermediate conformations or cell states for training downstream predictors (e.g., stability, binding affinity, trajectory inference).
- Sector(s): Software/ML for life sciences; Academia.
- Tools/Workflow: EntangledSBM-driven trajectory augmentation; label propagation along paths; curriculum training using interpolated states.
- Assumptions/Dependencies: Augmented samples reflect realistic dynamics (validated on held-out sets); avoid distributional shift; maintain uncertainty estimates.
- Multi-agent formation transition planning in simulation
- Description: Plan collision-aware formation changes for swarms by exploiting entangled velocity conditioning and target-directed bias under stochastic dynamics (simulation-level).
- Sector(s): Robotics (simulation), Defense/industrial R&D.
- Tools/Workflow: Simulation environments (e.g., Gazebo, Isaac Sim); define target formations as distributions; run EntangledSBM in silico for path proposals; handoff to deterministic controllers.
- Assumptions/Dependencies: Mapping domain dynamics to Langevin-style SDEs; stochastic-to-deterministic handoff; robustification for real hardware required.
- Teaching and methodological prototyping for stochastic control and Schrödinger bridges
- Description: Use EntangledSBM as a pedagogical and research scaffold to demonstrate stochastic optimal control, importance-weighted cross-entropy training, and path measures.
- Sector(s): Academia; Education.
- Tools/Workflow: Notebooks with toy systems and MD examples; comparisons of LV vs. CE objectives; replay buffer ablations.
- Assumptions/Dependencies: Availability of small, interpretable benchmarks; compute for hands-on labs.
Long-Term Applications
These applications are promising but require additional research, validation, scaling, or domain adaptation before deployment.
- Patient-specific therapy response prediction from scRNA-seq
- Description: Predict how an individual’s cell populations transition under candidate drugs/doses; support precision medicine.
- Sector(s): Healthcare/clinical decision support; Policy/regulatory science.
- Tools/Workflow: Patient scRNA-seq → manifold modeling → EntangledSBM trajectory forecasts per drug → rank and simulate combination schedules; clinical UI.
- Assumptions/Dependencies: Deep clinical validation; robust handling of patient heterogeneity and batch effects; interpretability and regulatory approval.
- Pathway-constrained de novo molecular design
- Description: Combine generative design with EntangledSBM to favor ligands that induce desired conformational transitions and path ensembles (not just endpoint binding).
- Sector(s): Pharma/biotech; Computational chemistry.
- Tools/Workflow: Multi-objective optimization with docking/scoring + path feasibility; iterative loop with wet-lab assays.
- Assumptions/Dependencies: Efficient end-to-end scoring of “path inducibility”; scalable integration with generative models; reliable free-energy estimates along paths.
- Closed-loop, autonomous perturbation design in high-throughput biology
- Description: Active learning system proposes perturbations (drugs/CRISPR/cytokines) predicted to drive cell clusters to target states; experiments feed back to retrain models.
- Sector(s): Biotech platforms; Pharma screening.
- Tools/Workflow: LIMS integration; robotic wet-lab; EntangledSBM-in-the-loop optimization; experiment scheduler.
- Assumptions/Dependencies: Safety and lab automation; robust uncertainty quantification; cost-effective iteration cycles.
- Real-time control of robotic swarms with entangled dynamics
- Description: Move from simulation to hardware by adapting EntangledSBM to low-latency, partially deterministic controllers with safety constraints.
- Sector(s): Robotics; Industrial automation.
- Tools/Workflow: Model predictive control that leverages learned bias proposals; on-board inference; safety shields.
- Assumptions/Dependencies: Bridging stochastic SDE controls to deterministic actuation; certification for safety-critical tasks.
- Traffic and crowd-flow interventions via distributional targets
- Description: Treat traffic/crowds as interacting agents; design interventions (signals, incentives) to shift distributions toward desired macrostates.
- Sector(s): Smart cities; Transportation; Public policy.
- Tools/Workflow: Agent-based simulators with stochastic dynamics; EntangledSBM for intervention design; digital twins.
- Assumptions/Dependencies: Realistic calibration; ethical considerations; non-stationarity and human behavior complexity.
- Market microstructure and multi-agent financial simulation
- Description: Model interacting agents with entangled dynamics to stress-test policies and liquidity interventions aimed at target market states.
- Sector(s): Finance; Risk management.
- Tools/Workflow: Synthetic order-book simulators; target distributions for volatility/liquidity; intervention policy search.
- Assumptions/Dependencies: Valid stochastic abstractions; data access; governance and compliance.
- Rare-event sampling in materials and energy systems
- Description: Efficiently sample phase transitions, defect dynamics, catalysis pathways by coupling DFT/ML potentials with EntangledSBM to produce path ensembles.
- Sector(s): Energy; Materials discovery.
- Tools/Workflow: ML interatomic potentials; CV discovery (e.g., TICA, VAMPnets); downstream free-energy methods.
- Assumptions/Dependencies: Accurate energy surfaces; scalable surrogates for large systems; high-performance computing.
- Cryo-EM flexible fitting and pathway reconstruction between conformational states
- Description: Use EM density–derived target distributions to guide trajectories that connect heterogeneous maps via physically plausible intermediates.
- Sector(s): Structural biology.
- Tools/Workflow: EM map likelihood as πB; EntangledSBM-guided morphing; validation with map cross-correlation.
- Assumptions/Dependencies: Robust target gradients from EM likelihoods; managing noise and resolution variability.
- Protein engineering via transition-state stabilization insights
- Description: Use path ensembles to identify residues stabilizing key intermediates/TS; propose mutations to tune kinetics or allostery.
- Sector(s): Biotechnology; Enzyme design.
- Tools/Workflow: Path clustering; ΔΔG predictions for mutants along paths; experimental kinetic assays.
- Assumptions/Dependencies: Reliable mapping from path features to functional outcomes; significant wet-lab validation.
- Regulatory pathway to reduce animal testing through validated in-silico forecasts
- Description: Standardize EntangledSBM-based protocols for MD and cell-state perturbation prediction as part of evidence packages.
- Sector(s): Policy/regulation; Pharma/biotech.
- Tools/Workflow: Benchmark suites; uncertainty calibration; submission-ready reports.
- Assumptions/Dependencies: Community standards; cross-lab reproducibility; acceptance criteria by regulators.
- Controlled generation in sequence models with interacting tokens
- Description: Adapt entangled SB ideas to sequence modeling to guide token interactions toward target sequence distributions (e.g., style/content constraints).
- Sector(s): Software/AI.
- Tools/Workflow: Score models for πB in sequence space; Transformer conditioning on target descriptors; off-policy training with replay.
- Assumptions/Dependencies: Well-defined target distribution over sequences; retooling SDE formalism for discrete tokens.
Notes on Feasibility and Key Assumptions (cross-cutting)
- Target specification: Many applications require a tractable target distribution πB and its gradient (or a reliable surrogate, e.g., KDE/score networks or indicator neighborhoods around a state).
- Dynamics validity: The Langevin/SDE abstraction should faithfully approximate domain dynamics; otherwise, domain-specific corrections or hybrid controllers are needed.
- Training stability and compute: Off-policy cross-entropy optimization with importance weights and replay buffers improves stability versus LV objectives but still needs careful tuning; scaling Transformers to large n requires memory-efficient attention or hierarchical architectures.
- Guarantees and step size: The non-increasing-distance property holds under small Δt and proper projection; violating these can degrade path quality.
- Reweighting for thermodynamics: When using EntangledSBM-generated paths for thermodynamic quantities, unbiased reweighting or enhanced sampling remains necessary.
- Generalization: The paper shows generalization to unseen targets via conditioning; robustness across broader shifts depends on target-feature design and data coverage.
Glossary
- Annealed temperature schedule: A technique where the simulation temperature is gradually changed (often decreased) during dynamics to help explore or stabilize states. "simulate the position and velocity using OpenMM \citep{eastman2017openmm} with an annealed temperature schedule."
- Bias force: An additional control term added to the dynamics to tilt the path distribution toward desired endpoints or distributions. "Schrödinger Bridge Matching (SBM) aims to parameterize a control or bias force "
- Brownian motion: A stochastic process modeling random fluctuations, used as the driving noise in SDEs. " is standard Brownian motion."
- Cross-Entropy objective: A convex objective over path measures that aligns a biased path distribution with a target bridge distribution via KL minimization. "we propose a cross-entropy objective that is convex with respect to the biased path measure "
- Dihedral angles: The torsion angles (e.g., backbone φ, ψ) that describe rotations around bonds in molecules and define conformational states. "defined by the backbone dihedral angles "
- Entangled bias force: A control force whose value for each particle depends on the positions and velocities of all other particles, capturing dynamic interactions. "an entangled bias force capturing the dynamic interactions between particles."
- Entangled Schrödinger Bridge (EntangledSB): The formulated problem of finding bias forces that couple multi-particle dynamics to transport an initial distribution to a target distribution. "We define the Entangled Schrödinger Bridge (EntangledSB) problem"
- Entangled Schrödinger Bridge Matching (EntangledSBM): The proposed framework that parameterizes and learns entangled bias forces to simulate interacting multi-particle trajectories. "we introduce Entangled Schrödinger Bridge Matching (EntangledSBM), a framework"
- Friction coefficient: A parameter in Langevin dynamics controlling damping due to the environment. " is the friction coefficient"
- Importance weight: A likelihood ratio used for off-policy optimization to reweight trajectories from a reference path measure to the target bridge measure. "we define an importance weight $w^\star(\boldsymbol{X}_{0:T}):=\frac{\mathrm{d}\mathbb{P}^\star}{\mathrm{d}\mathbb{P}^{v}(\boldsymbol{X}_{0:T})$"
- Kabsch algorithm: A method to find the optimal rotation that aligns two sets of coordinates, preserving distances. "using the Kabsch algorithm \citep{kabsch1976solution}"
- Kullback–Leibler divergence (KL-divergence): A measure of difference between distributions; here used to align biased path measures with the bridge distribution. "minimizes the KL-divergence from the bridge path distribution "
- Langevin dynamics: Stochastic dynamics that model motion under conservative forces with thermal noise and damping. "can be modelled with Langevin dynamics"
- Log-Variance divergence (LV divergence): A variational objective based on the variance of log-likelihood ratios between path measures. "we can adapt the log-variance (LV) divergence"
- Maximum mean discrepancy (RBF-MMD): A kernel-based distance between distributions used to assess reconstruction accuracy. "using the maximum mean discrepancy (RBF-MMD)"
- Mean-field assumption: An approximation where each particle interacts with the average effect of others, treating particles as exchangeable. "rely on the mean-field assumption, where all particles are exchangeable and interact only through averaged effects"
- Molecular dynamics (MD): Simulation of atomistic systems using physics-based dynamics to paper time evolution and interactions. "molecular dynamics (MD) simulation"
- Overdamped Langevin dynamics: A regime where inertia is negligible, leading to first-order position dynamics with noise. "overdamped Langevin dynamics, where inertia is negligible"
- Path integral: A formulation expressing objectives or probabilities as integrals over trajectories in path space. "can be expressed in path-integral form as"
- Path measure: A probability measure over entire trajectories induced by a given stochastic dynamics (control) law. "path measure generated with an arbitrary control "
- Potential energy landscape: The mapping from configurations to potential energies that governs forces and feasible pathways. "The velocities follow the potential energy landscape defined over the joint coordinates of the system "
- Replay buffer: A memory of previously simulated trajectories and associated weights used for off-policy training. "maintaining a replay buffer "
- Schrödinger Bridge Matching (SBM): A class of methods that learn control forces to match trajectories between prescribed endpoint distributions. "Schrödinger Bridge Matching (SBM) has enabled significant progress"
- Stochastic differential equation (SDE): An equation describing dynamics with deterministic and stochastic components, e.g., Langevin motion. "stochastic differential equations (SDEs) defined as"
- Stochastic optimal control (SOC): The theory and optimization of control inputs to stochastic systems to optimize expected rewards or costs. "stochastic optimal control (SOC) objective"
- Terminal reward: The scalar reward function evaluated at the final state of a trajectory, often the log-likelihood under the target distribution. "is the terminal reward that measures the log-probability under the target distribution."
- Time-lagged independent component analysis (TICA): A dimensionality reduction method that extracts slow collective variables from time-series (MD) data. "top two TICA components"
- Transition path sampling (TPS): Methods for generating trajectories between metastable states, focusing on rare event transitions. "transition path sampling (TPS)"
- Underdamped Langevin dynamics: A regime where inertia is significant, leading to second-order dynamics in positions and velocities. "underdamped Langevin dynamics with small "
- Wasserstein distance: An optimal transport-based metric between probability distributions, here used to compare simulated and target cell states. "Wasserstein distances ( and )"
Collections
Sign up for free to add this paper to one or more collections.