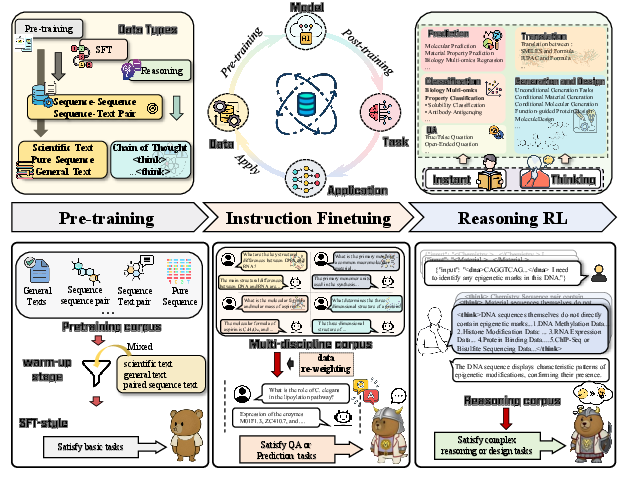
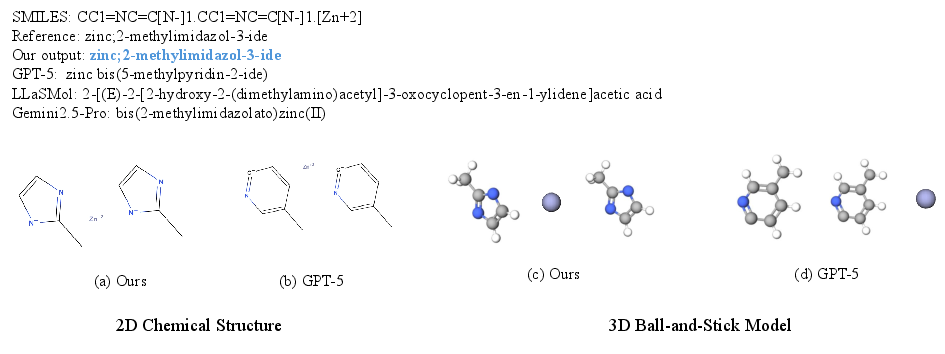
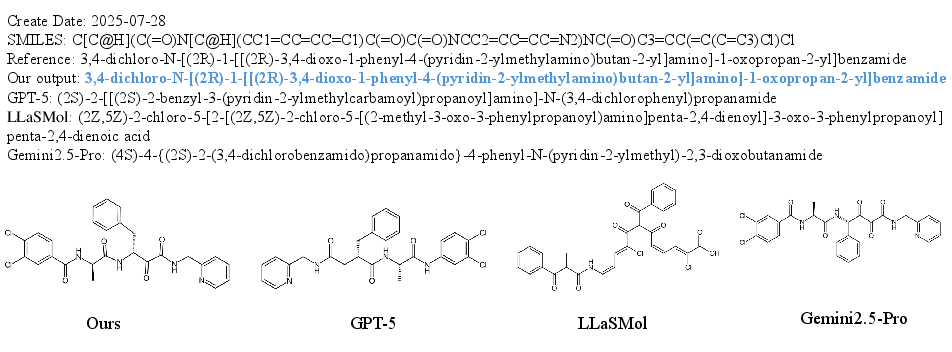
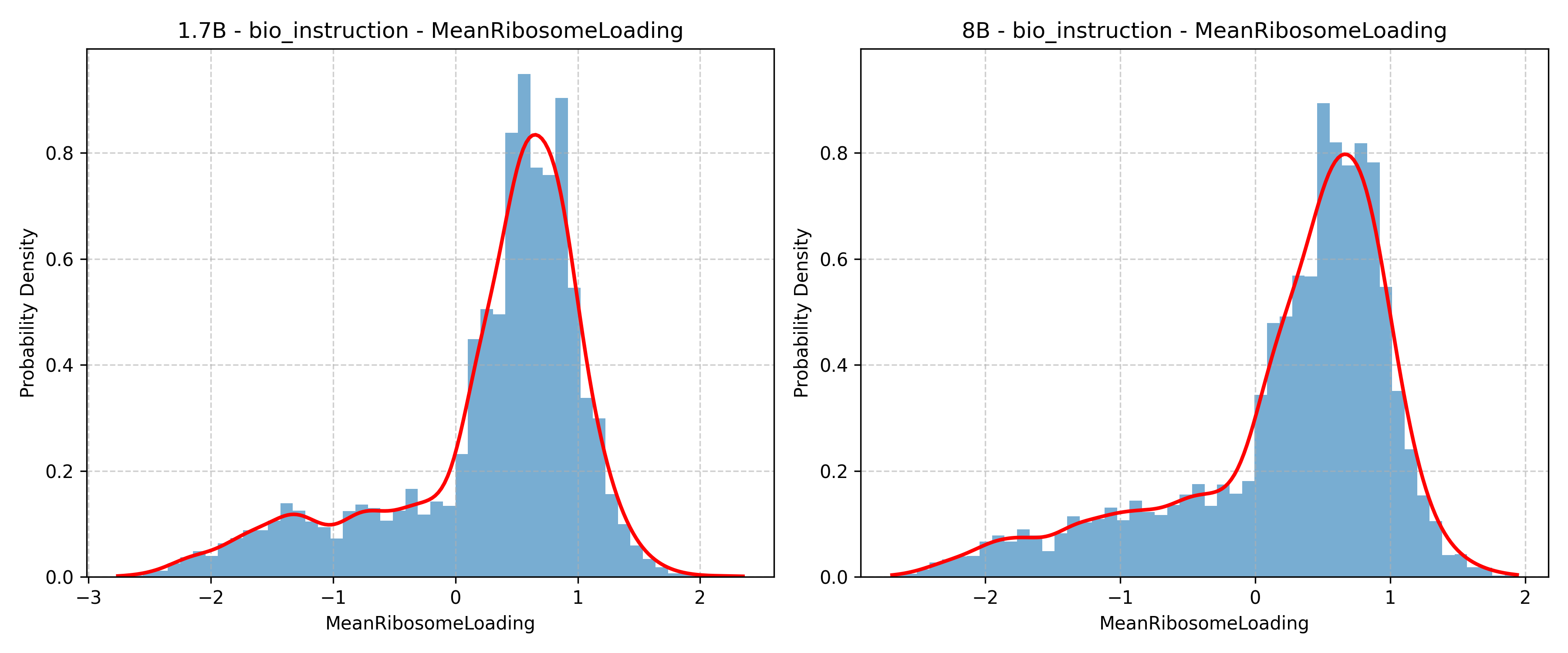
SciReasoner: Laying the Scientific Reasoning Ground Across Disciplines (2509.21320v1)
Abstract: We present a scientific reasoning foundation model that aligns natural language with heterogeneous scientific representations. The model is pretrained on a 206B-token corpus spanning scientific text, pure sequences, and sequence-text pairs, then aligned via SFT on 40M instructions, annealed cold-start bootstrapping to elicit long-form chain-of-thought, and reinforcement learning with task-specific reward shaping, which instills deliberate scientific reasoning. It supports four capability families, covering up to 103 tasks across workflows: (i) faithful translation between text and scientific formats, (ii) text/knowledge extraction, (iii) property prediction, (iv) property classification, (v) unconditional and conditional sequence generation and design. Compared with specialist systems, our approach broadens instruction coverage, improves cross-domain generalization, and enhances fidelity. We detail data curation and training and show that cross-discipline learning strengthens transfer and downstream reliability. The model, instruct tuning datasets and the evaluation code are open-sourced at https://huggingface.co/SciReason and https://github.com/open-sciencelab/SciReason.
Paper Prompts
Sign up for free to create and run prompts on this paper using GPT-5.
Top Community Prompts
Explain it Like I'm 14
What is this paper about?
This paper introduces SciReasoner, a large AI model designed to “think like a scientist.” It can read and write normal text and also understand special scientific formats used in chemistry, biology, and materials science—things like DNA letters, protein sequences, and molecule codes. The goal is to help with many kinds of science tasks, from translating between formats to predicting properties and even designing new molecules or proteins.
What questions were the researchers asking?
In simple terms, they wanted to know:
- Can one AI model learn to handle many different science fields (chemistry, biology, materials) instead of building a separate model for each?
- Can it decide when to give a quick answer and when to show step‑by‑step reasoning, like a student who sometimes does mental math and sometimes shows the full working?
- Can training the model to “think out loud” (show its steps) make it more accurate and reliable on tough scientific tasks?
- Can we check and improve the model’s answers using fair scoring methods that give partial credit (not just right/wrong), so it learns stably and improves over time?
How did they build and train the model?
They trained SciReasoner in three main stages, using ideas that are easy to picture:
1) Pretraining: teaching the basics at scale
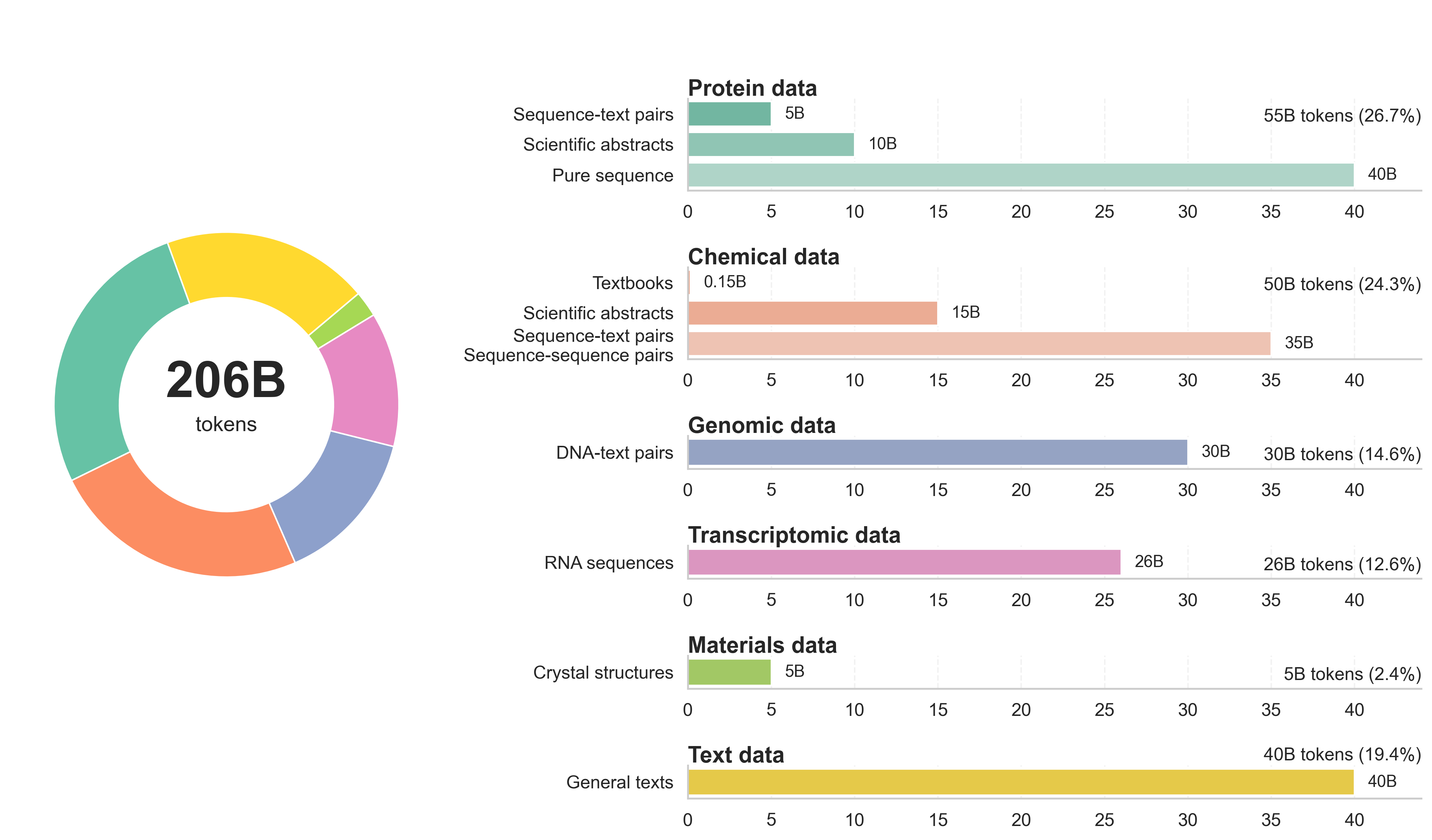
They first taught the model general science “languages” by feeding it a huge mix of data—about 206 billion tokens (tiny chunks of text or symbols). This included:
- Scientific text (like articles and descriptions)
- Pure sequences (DNA/RNA letters, protein amino acids, and molecule strings such as SMILES/IUPAC/SELFIES)
- Pairs of sequences and text (so it learns to connect formats with plain language)
- General text (to keep normal conversation skills)
Think of this as teaching the model many alphabets and grammars so it becomes fluent in both human language and scientific “code.”
2) Supervised fine-tuning (SFT): practicing real tasks
Next, they gave the model about 40 million example tasks and answers, spanning roughly 100 different scientific tasks. These included:
- Translation between formats (e.g., SMILES ↔ IUPAC names)
- Finding facts in text (knowledge extraction)
- Predicting numbers (like solubility or stability)
- Classifying properties (e.g., “toxic or not,” “stable or not”)
- Generating sequences/designs (like new molecules/proteins under constraints)
They “tagged” sequences with markers like <dna>…</dna> or <SMILES>…</SMILES> so the model knows exactly what it’s looking at.
3) Reasoning training: teaching when and how to think
They added two important steps so the model learns to reason well:
- Adaptive thinking (Instant vs. Thinking tasks):
- Instant tasks: keep short, direct answers.
- Thinking tasks: replace training examples with “chain‑of‑thought” (CoT), which are step‑by‑step rationales ending with the final answer.
- This helps the model save time on simple problems and think carefully on hard ones.
- Reinforcement learning (RL) with smarter rewards:
- They grouped rewards by task type:
- Distance-based: “How close is your number to the truth?”
- Matching-based: “Does your extraction/translation match the reference?”
- Tool-verified: “Do scientific tools say your answer is valid?” (like checking chemistry with pro software)
- Reward softening: Instead of only right (1) or wrong (0), they give partial credit on a 0–1 scale (like grading with partial points), which makes learning smoother and more reliable.
- Mid‑difficulty focus: They train more on questions the model sometimes gets right and sometimes wrong (not the super easy or the impossible ones), which is where learning happens fastest.
What did they find, and why does it matter?
They report that a single, unified model can:
- Cover a very wide set of scientific tasks (up to 103 tasks across chemistry, biology, and materials).
- Achieve state-of-the-art results on 54 tasks and place in the top 2 on 101 tasks.
- Translate faithfully between text and scientific formats, extract knowledge from papers, predict properties, classify properties, and design sequences under constraints.
- Generalize across fields better than many specialist systems (it reuses what it learns in one area to help in another).
- Decide when to think step-by-step and when to answer directly, improving both speed and accuracy.
Why it matters:
- Scientists often juggle many tools and formats. A model that speaks all these “languages” can save time and reduce errors.
- Showing reasoning steps (when needed) makes results easier to check, which builds trust.
- Better cross‑domain learning means fewer separate systems and less re‑training when new tasks come up.
What could this change in the future?
Here’s what this research could enable:
- Faster discovery: Quickly screen or design molecules, proteins, or materials with specific targets (e.g., stability, safety, or synthesizability).
- Smoother workflows: Translate between human-friendly text and machine-friendly scientific code without losing meaning.
- More reliable AI science helpers: Step‑by‑step reasoning, tool‑checked answers, and partial‑credit learning can reduce mistakes and make outputs easier to verify.
- Broad, shareable tools: Because the model, datasets, and evaluation code are open-sourced, researchers and students can build on this work for many scientific areas.
In short, SciReasoner is like a multilingual science student who knows when to think out loud and when to answer directly, can read lab notes and complex codes, and learns from fair feedback. It brings many scientific skills into one model, aiming to make research faster, clearer, and more connected across disciplines.
Knowledge Gaps
Knowledge gaps, limitations, and open questions
Below is a single, focused list of concrete gaps and unresolved questions that could guide future research on SciReasoner.
- Data transparency and contamination safeguards: The paper does not detail dataset licensing, deduplication, split strategy (especially across pretraining/SFT/evaluation), or contamination auditing—particularly crucial given PubMed/PubChem/C4 usage and overlapping benchmark tasks. Provide reproducible scripts and leakage checks that verify no test items (or close paraphrases) were present in pretraining/SFT.
- Modality coverage beyond text/sequences: The model appears centered on natural language and 1D sequences. Integration of 3D structures (proteins/materials), graphs, spectra (NMR/MS/IR), microscopy images, and time-series experimental data is not addressed. Evaluate whether adding structured modalities improves property prediction and design; define tokenization/encoders for CIF/graph/surface representations.
- Materials input fidelity: Materials pretraining relies on LLM-generated natural-language templates from CIF and metadata, but direct ingestion of CIF or graph-based crystalline structures and round-trip translation (text ↔ CIF) are not shown. Test direct structural parsing, equivalence-preserving translation, and property prediction with physically grounded inputs.
- Long-context limits: With 8192-token truncation, the model may be unable to handle full papers, protocols, multi-sample experiment logs, or genomic regions requiring long-range context. Benchmark long-context tasks (e.g., literature QA over multi-section PDFs, whole-transcriptome analyses) and paper scaling of context length and retrieval augmentation.
- Instant vs. thinking task assignment: Criteria for classifying tasks as “instant” vs “thinking” are not specified or validated. Provide an automatic, data-driven assignment (e.g., via entropy/solve-rate/format compliance) and sensitivity analyses showing how misclassification affects performance and CoT quality.
- Chain-of-thought faithfulness: CoT correctness is judged by final-answer alignment or a GPT-3.5 judge; the paper does not assess whether rationales are causally faithful to the model’s decision process or scientifically valid. Introduce faithfulness audits (e.g., input perturbation tests, tool-grounded derivation checks, rationale consistency with physics/stoichiometry) and human expert evaluation.
- RL reward calibration function g(·): The “soft reward” mapping to [0,1] is unspecified (functional form, monotonicity, per-task scaling). Publish the exact calibration functions, rationale, and cross-task comparability analyses; run ablations to show sensitivity of training stability and final performance to reward scaling.
- Scientific tool-verified rewards: The paper mentions tool-verified rewards but does not enumerate tools, versions, validation protocols, or failure-handling (e.g., RDKit canonicalization settings, protein scoring tools, materials simulators). Provide a catalog of tools, their configurations, and robustness tests to parser/tool errors.
- Mid-difficulty RL filtering thresholds: The empirical solve-rate band (0.125–0.875) and K=1000 sampling are heuristic. Evaluate alternative bands, adaptive per-task thresholds, and curriculum strategies to optimize policy improvement and avoid biasing toward mid-range difficulty tasks.
- Dynamic temperature tuning for rollouts: Tuning is described qualitatively; exact criteria (diversity metrics, format compliance thresholds), search procedure, and stability are not provided. Standardize and release the tuning protocol, including metrics and stopping rules.
- Ablation studies: The report lacks ablations isolating contributions from (i) pretraining mixture (sequence/text proportions), (ii) SFT re-weighting, (iii) annealed cold-start replacement vs mixing, and (iv) RL with vs without soft rewards and tool verification. Provide controlled ablations with statistical testing across representative tasks.
- Statistical rigor and uncertainty: Claims of SOTA/top-2 performance are not accompanied by confidence intervals, multiple seeds, or significance tests. Add variance estimates, seed robustness, and significance testing for key benchmarks.
- Baseline comparability: Closed-source baselines were evaluated on 1,000-sample subsets for large test sets; it is unclear if sampling was stratified or matched to task difficulty and distribution. Describe sampling procedures, ensure consistent subsets across models, and report full-set comparisons where feasible.
- Data imbalance and re-weighting: While data re-weighting is mentioned, the method (weights, criteria, schedule) and its quantitative impact are not detailed. Provide the weighting scheme, diagnostics (per-task loss/accuracy), and guidelines for preventing sub-task degradation in mixed training.
- Tokenization details: “Task-aware tokenization” is referenced, but token definitions for scientific sequences (e.g., amino acid/nucleotide alphabets, SMILES grammar tokens, special tags) and normalization/canonicalization rules are not provided. Release tokenizer specs and analyze their effects on cross-representation translation fidelity.
- Property metrics and equivalence checking: For translation tasks (e.g., IUPAC ↔ SMILES ↔ formula), the canonicalization/equivalence criteria are not fully specified (tautomer handling, salts/mixtures, stereochemistry). Define standardized equivalence checks and report failure modes (e.g., stereochemistry loss, invalid valences).
- Safety and dual-use risk: The paper does not propose safeguards for generating hazardous molecules/proteins/materials (toxicity, biosecurity) or medical misinformation. Develop safety filters, risk classifiers, red-teaming evaluations, and publication policies for constrained generation.
- Real-world validation: There is no experimental or high-fidelity simulation validation of generated designs or predicted properties (e.g., synthesizability, stability, binding). Partner with labs to validate a subset of outputs and report hit rates, synthesis success, and physical constraint adherence.
- Generalization to unseen tasks/modalities: Although cross-domain generalization is claimed, zero-shot transfer to truly novel tasks or representations (not present in SFT) is not systematically tested. Construct compositional/novel-task benchmarks and measure out-of-distribution robustness.
- Hallucination and factuality auditing: Beyond correctness on auto-gradable tasks, the paper lacks a comprehensive hallucination audit for literature QA and entity extraction. Quantify hallucination rates, cite-resolution accuracy, and implement provenance tracking (e.g., citation linking to source passages).
- Multilingual scientific text: The corpus appears predominantly English; multilingual capability for non-English scientific literature is not evaluated. Assess performance across major scientific languages and consider multilingual pretraining augmentation.
- Lengthy training inconsistencies: The SFT data distribution table reports totals that conflict with narrative numbers (e.g., “39.824M” vs “63.35M”). Clarify actual counts, splits, and any filtering applied; ensure internal consistency across tables.
- Compute/resource reporting: Training uses 128–256 A800 GPUs, but energy cost, efficiency, and scaling-law analyses are missing. Provide compute budgets, training time, efficiency metrics, and parameter-scaling experiments beyond 8B to inform reproducibility and sustainability.
- External-judge dependency: Use of GPT-3.5-turbo for semantic adjudication introduces a closed-model dependency and potential bias. Evaluate open-source judges, inter-annotator agreement with human experts, and robustness of results to judge selection.
- Ease of adding new tasks: The paper claims broad task coverage, but the workflow to add new scientific tasks (schemas, tags, rewards, tools) is not documented. Provide a modular template, data-to-reward pipeline, and case studies of onboarding a new task end-to-end.
- Privacy and compliance: Handling of potentially sensitive biomedical data (e.g., human-derived sequences) and compliance with database licenses is not discussed. Document privacy safeguards, consent/usage constraints, and license compliance procedures.
- Failure mode catalog: A systematic error analysis (format non-compliance, invalid chemistry, biologically implausible sequences, materials inconsistency) is missing. Publish a taxonomy of errors, quantitative rates per domain, and mitigation strategies.
Practical Applications
Immediate Applications
The following applications can be deployed with the model’s current capabilities, datasets, and evaluation code, especially when paired with existing domain tools (e.g., RDKit, SELFIES, BLAST, CIF parsers) and human oversight where needed.
- Bold cross-format chemical translation in ELNs/LIMS
- Sectors: healthcare (R&D), pharmaceuticals, chemicals, software
- Use case: Seamless SMILES ↔ IUPAC ↔ formula conversion embedded in electronic lab notebooks (ELNs), LIMS, and procurement systems to unify inventory, SDS linking, and compliance workflows.
- Tools/products/workflows: RDKit-powered “Molecular Translator” microservice; ELN/LIMS plugin; batch conversion pipelines; validation with canonicalization.
- Assumptions/dependencies: Ambiguous nomenclature requires disambiguation; canonical equivalence checks; human review for edge cases; consistent tagging (e.g., <SMILES>...</SMILES>).
- Literature mining and knowledge graph construction
- Sectors: academia, biotech, pharma, materials, policy
- Use case: Automatic chemical/protein/material entity recognition, relation extraction (e.g., protein–ligand, reaction steps), and Q&A from literature to build living knowledge graphs and speed systematic reviews.
- Tools/products/workflows: “Literature Miner” with RAG, MeSH/ChEBI/UMLS ontologies; provenance tracking; queryable graph database (e.g., Neo4j).
- Assumptions/dependencies: Requires ontology alignment; rigorous fact-check via domain tools; transparent citations; guardrails against hallucinations.
- Rapid property screening (non-clinical)
- Sectors: pharma (early discovery), materials, energy, agriculture
- Use case: High-throughput triage for solubility, toxicity flags, bandgap, stability, or other properties to prioritize candidates before expensive simulations/experiments.
- Tools/products/workflows: SciReasoner + RDKit + Materials Project/JARVIS APIs; uncertainty scoring; calibration curves; “screen-and-rank” dashboards.
- Assumptions/dependencies: Not for clinical decision-making; external validation required; domain-specific tolerances; continuous reward–calibrated metrics (e.g., RMSE-to-[0,1] mapping).
- Constraint-aware sequence ideation (protein/RNA)
- Sectors: academia, biotech (research), education
- Use case: Generate protein or RNA sequences satisfying basic constraints (length, motif inclusion, synthesis feasibility) for hypothesis generation and classroom labs.
- Tools/products/workflows: Sequence generation interface; pre-checks with BLAST/CD-HIT; synthesis feasibility heuristics; design-notebook export.
- Assumptions/dependencies: Wet-lab validation needed; biosafety compliance; conservative deployment in low-risk contexts; sequence tags (<protein>, <rna>) for accurate parsing.
- Retrosynthesis sketching and reaction pathway assistance
- Sectors: chemicals, pharma, CROs
- Use case: Propose plausible reaction steps, reagents, and conditions based on USPTO data and literature; link to vendor catalogs and risk flags.
- Tools/products/workflows: Reaction-rule engines (e.g., RDKit reaction SMARTS); “solve–check” stoichiometry validator; SDS integration.
- Assumptions/dependencies: Chemist oversight mandatory; safety/regulatory constraints; tool-verified reward functions to penalize violations (e.g., unbalanced equations).
- CIF-to-plain-language structure narrations
- Sectors: materials science, energy, manufacturing
- Use case: Convert crystallographic information files and annotations into human-readable summaries for lab notes, reports, and teaching.
- Tools/products/workflows: CIF parsers; “Structure Narrator” service; space-group/coordination summaries with uncertainty labels.
- Assumptions/dependencies: Accurate parsing of CIFs; cross-referencing with databases (Materials Project, GNoME).
- Scientific data quality control and tagging
- Sectors: software, academia, data platforms
- Use case: Automated detection of malformed SMILES/SELFIES, tag missing sequence metadata, normalize representations across datasets.
- Tools/products/workflows: QC pipeline with RDKit/SELFIES validators; tag-enforcement transformers; dataset health dashboards.
- Assumptions/dependencies: Rule-based + LLM checks; conservative discard policies; audit trails.
- Educational scientific reasoning tutor
- Sectors: education (secondary, tertiary), public outreach
- Use case: Teach chemical nomenclature, structural reasoning, protein function summaries, and step-by-step problem-solving with adaptive “instant vs thinking” modes.
- Tools/products/workflows: Classroom chat assistants; assignment feedback with CoT rationales; curated curriculum modules.
- Assumptions/dependencies: Instructor oversight; alignment with standards; guardrails for safety-sensitive topics.
- “Solve–check” compliance utilities for unit and stoichiometry consistency
- Sectors: education, software, chemicals
- Use case: Automated unit conversions, stoichiometry balance checks, and constraint verification during lab prep and reporting.
- Tools/products/workflows: Python SDK; notebook extensions; CLI validation pipelines.
- Assumptions/dependencies: Domain tool integration; standardized units; overridable checks for edge cases.
- Cross-domain research copilots in notebooks
- Sectors: academia, R&D
- Use case: In-notebook assistant that translates formats, runs extraction, predicts properties, and drafts methods sections with references and device-ready sequence tags.
- Tools/products/workflows: Jupyter/lab plugin; local RAG; action toolchain with RDKit/BLAST/CIF parsers.
- Assumptions/dependencies: Compute availability; dataset licensing; explicit provenance and versioning.
- Patent reaction analytics
- Sectors: IP/legal, pharma
- Use case: Extract reaction classes and conditions from patent corpora (e.g., USPTO), cluster innovations, and monitor competitive trends.
- Tools/products/workflows: Patent ingestion pipeline; reaction classification models; dashboards for counsel and strategy.
- Assumptions/dependencies: Licensing/compliance; ambiguity handling; expert review for legal contexts.
- EHS/ESG chemical inventory harmonization
- Sectors: enterprise safety/compliance, manufacturing
- Use case: Map inventory entries from varied formats to standardized identifiers, auto-link SDS/hazard classes, and flag restricted substances.
- Tools/products/workflows: Inventory translators; SDS linkage; compliance reports.
- Assumptions/dependencies: Up-to-date hazard databases; exact-match thresholds; auditability.
- Supplier/spec sheet normalization
- Sectors: manufacturing, procurement
- Use case: Convert heterogeneous spec sheets to machine-readable forms, align with internal schemas, and perform consistency checks across vendors.
- Tools/products/workflows: Spec parser; schema mapper; QA dashboards.
- Assumptions/dependencies: Schema agreement; exception-handling; human-in-the-loop QA.
- Benchmarking and reproducible evaluation suites
- Sectors: academia, ML tooling
- Use case: Adopt open-source evaluation code and datasets to benchmark in-house models, reproduce tasks, and perform ablation studies.
- Tools/products/workflows: Standardized task runners; CI-integrated benchmarks; leaderboard hosting.
- Assumptions/dependencies: Hardware availability; consistent tokenization/tagging; well-defined task metrics.
Long-Term Applications
These applications require scaling, additional research, integration with robotics or clinical workflows, regulatory approvals, robust uncertainty quantification, and broader community/standards adoption.
- Closed-loop autonomous discovery with robotic labs
- Sectors: biotech, materials, chemistry
- Use case: End-to-end “read–reason–design–execute” loops where the model proposes experiments, robots run them, and reward-shaped RL updates the policy.
- Tools/products/workflows: Robotic lab orchestration; active learning; tool-verified rewards; lab LIMS integration.
- Assumptions/dependencies: Reliable uncertainty estimation; biosafety and chemical safety controls; experiment tracking; fail-safe policies.
- AI copilot for drug discovery (design–make–test–analyze)
- Sectors: pharma, biotech
- Use case: Multi-objective molecular design optimizing ADMET, synthesis feasibility, and patent landscape with explicit chain-of-thought rationales.
- Tools/products/workflows: Docking/MD coupling; synthesis planning; patent RAG; portfolio dashboards.
- Assumptions/dependencies: Preclinical-only until validated; rigorous external benchmarks; regulatory pathways; governance for generative bio risk.
- Clinical decision support for lab interpretation (research-phase to regulated use)
- Sectors: healthcare
- Use case: Literature-grounded interpretation of lab results, assay selection suggestions, and protocol rationales, evolving toward regulated CDS as evidence accrues.
- Tools/products/workflows: EHR-integrated modules; guideline alignment; audit trails.
- Assumptions/dependencies: Prospective validation; bias audits; regulatory clearance; data privacy.
- Advanced protein/antibody engineering platforms
- Sectors: biotech, industrial enzymes
- Use case: Generative design with stability, specificity, immunogenicity, and manufacturability objectives; iterative wet-lab feedback.
- Tools/products/workflows: “Antibody Studio”; sequence–structure co-modeling; manufacturability simulators.
- Assumptions/dependencies: Robust experimental pipelines; IP considerations; ethical safeguards.
- Materials discovery for energy (batteries, catalysis, photovoltaics)
- Sectors: energy, materials, manufacturing
- Use case: Propose novel compositions and structures subject to stability and performance constraints; verify via DFT/HPC and experiments.
- Tools/products/workflows: DFT integration (JARVIS, SNUMAT); high-throughput screening; lab automation.
- Assumptions/dependencies: Compute budgets; uncertainty-aware ranking; lab capacity for synthesis/testing.
- Programmable synthetic biology circuits (RNA switches, CRISPR guides)
- Sectors: synthetic biology, agriculture, biomanufacturing
- Use case: Co-design genetic parts under biosafety and off-target constraints; iterative learning from wet-lab results.
- Tools/products/workflows: gRNA off-target prediction; pathway optimization; biosafety reward shaping.
- Assumptions/dependencies: Regulatory approvals; environmental risk assessments; containment policies.
- Government evidence synthesis and horizon scanning
- Sectors: policy, public health, environmental agencies
- Use case: Continual mining of scientific literature and patents to identify emerging risks/opportunities with transparent reasoning trails.
- Tools/products/workflows: Public dashboards; expert review workflows; SAR-based policy memos.
- Assumptions/dependencies: Independent audits; bias mitigation; accountability frameworks; procurement/FOIA compliance.
- Community standards for multimodal scientific tagging and interchange
- Sectors: standards bodies, publishers, repositories
- Use case: Adopt task-aware tokenization and tags (<dna>, <rna>, <protein>, <SMILES>) as community standards for reproducible AI pipelines and data interoperability.
- Tools/products/workflows: Schema registries; validator toolkits; publisher submission checks.
- Assumptions/dependencies: Broad stakeholder adoption; backward compatibility; governance.
- Federated, provenance-rich “SciGraph” knowledge bases
- Sectors: academia, industry consortia
- Use case: Multi-institution graph of entities, relations, and claims with confidence and source trails, powering discovery and meta-analyses.
- Tools/products/workflows: Federated ingestion; confidence calibration; conflict resolution protocols.
- Assumptions/dependencies: Data-sharing agreements; privacy/security controls; citable provenance.
- Adaptive pedagogy grounded in “instant vs thinking” modes
- Sectors: education, edtech
- Use case: Curricula that train students when to reason step-by-step vs deliver concise answers; automated feedback on scientific method and experimental design.
- Tools/products/workflows: LMS integration; rubric-aligned CoT; formative assessment analytics.
- Assumptions/dependencies: Teacher training; equitable access; age-appropriate guardrails.
- Generative bio safety gating and compliance tooling
- Sectors: platforms, biosafety committees
- Use case: Reward-shaped filters that block high-risk designs and flag sensitive queries; compliance logs for institutional oversight.
- Tools/products/workflows: Red-teaming harnesses; safety classifiers; audit dashboards.
- Assumptions/dependencies: Shared risk taxonomies; periodic updates; human escalation paths.
- Environmental remediation design (enzymes, materials)
- Sectors: environment, municipal services, NGOs
- Use case: Suggest biocatalysts or materials for pollutant capture/breakdown; simulation-to-field pipeline with uncertainty management.
- Tools/products/workflows: Kinetics models; pilot field trials; regulatory reporting.
- Assumptions/dependencies: Field validation; ecological assessments; community engagement.
- Marketplace of composable scientific workflows
- Sectors: software ecosystems, CROs
- Use case: Publish and monetize modular “capability families” (translation, extraction, prediction, design) as interoperable microservices with tool-verified rewards.
- Tools/products/workflows: API gateway; workflow orchestrator; billing/quotas; reproducibility badges.
- Assumptions/dependencies: Security/hardening; IP management; service-level guarantees.
Cross-cutting assumptions and dependencies
- Reliability and verification: Model outputs require tool-verified checks (stoichiometry, unit consistency, canonical structure equivalence), uncertainty quantification, and domain-expert review for high-stakes decisions.
- Data quality and coverage: Performance depends on the breadth and cleanliness of pretraining/SFT corpora; task re-weighting and calibration are necessary for imbalanced domains.
- Safety and compliance: Biosafety, chemical safety, and responsible use policies must be embedded; regulatory approvals are essential for clinical or environmental deployment.
- Integration and scalability: Success hinges on robust APIs, workflow orchestration, hardware acceleration, and compatibility with existing scientific tooling and databases.
- Governance and provenance: Transparent chain-of-thought, citations, dataset licensing, and audit trails are critical for trust, reproducibility, and policy adoption.
Glossary
- Annealed cold-start (ACS): A training strategy that gradually adapts the model to produce chain-of-thought on tasks needing stepwise reasoning while preserving direct answers where appropriate. "Before reinforcement learning, we apply an annealed cold-start (ACS) adaptation"
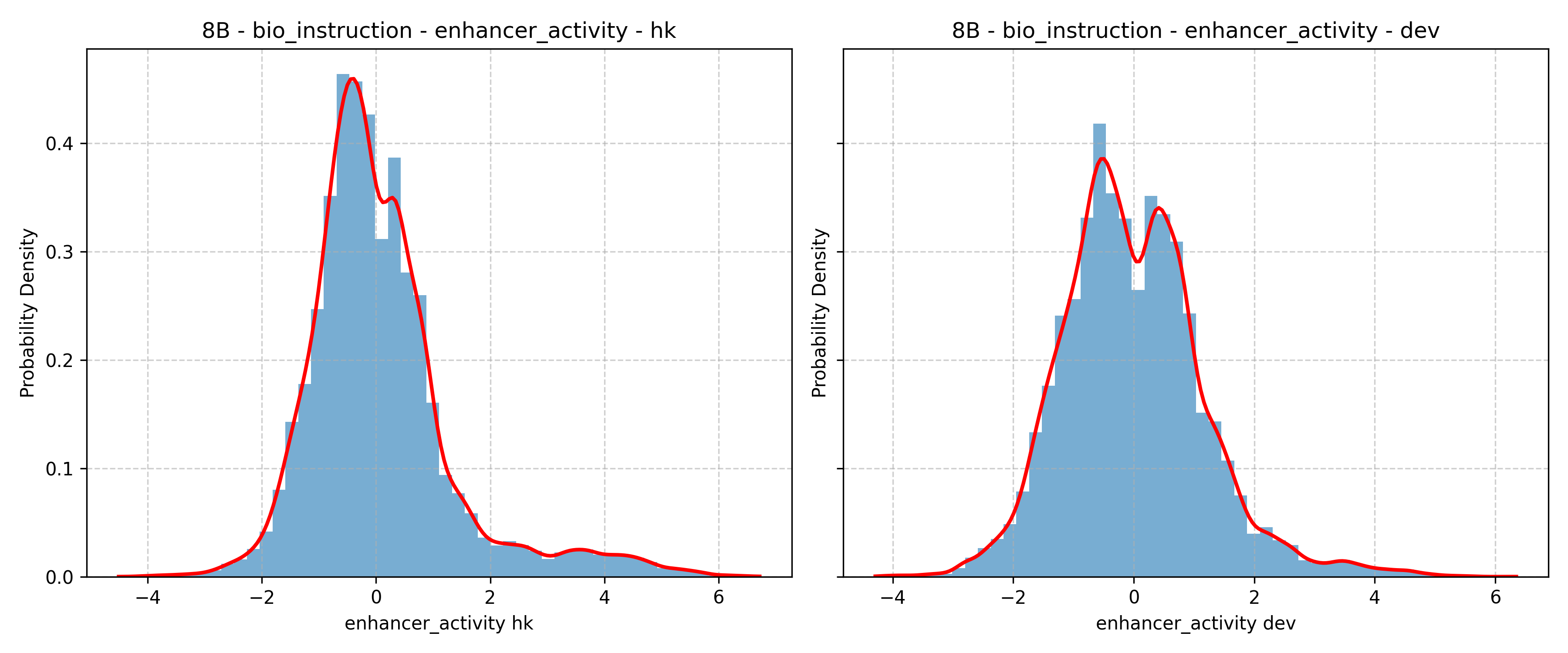
- Band gap: The energy difference between the valence and conduction bands in a material, critical for determining electronic properties. "electronic and mechanical properties (band gap, density, elastic moduli)"
- BEACON: A biology-focused dataset/benchmark used to train or evaluate sequence-based models. "BEACON~\cite{ren2024beacon}"
- BERTScore: A reference-based metric that uses contextual embeddings to measure semantic similarity between texts. "language-based scoring metrics (e.g., BERTScore)"
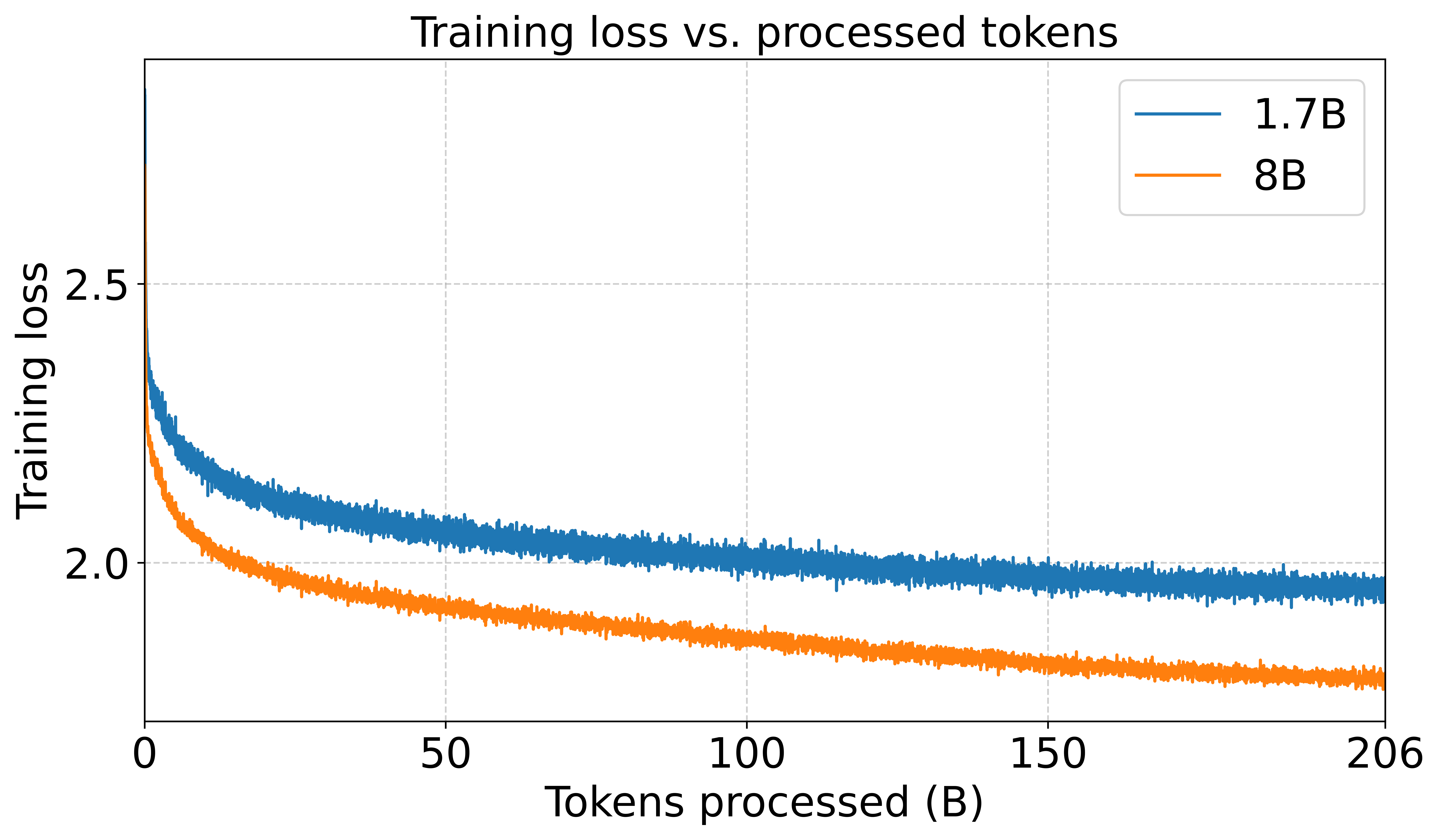
- bfloat16 mixed-precision: A 16-bit floating-point format used to reduce memory and accelerate training with minimal loss in numerical stability. "We train the model for one epoch using bfloat16 mixed-precision."
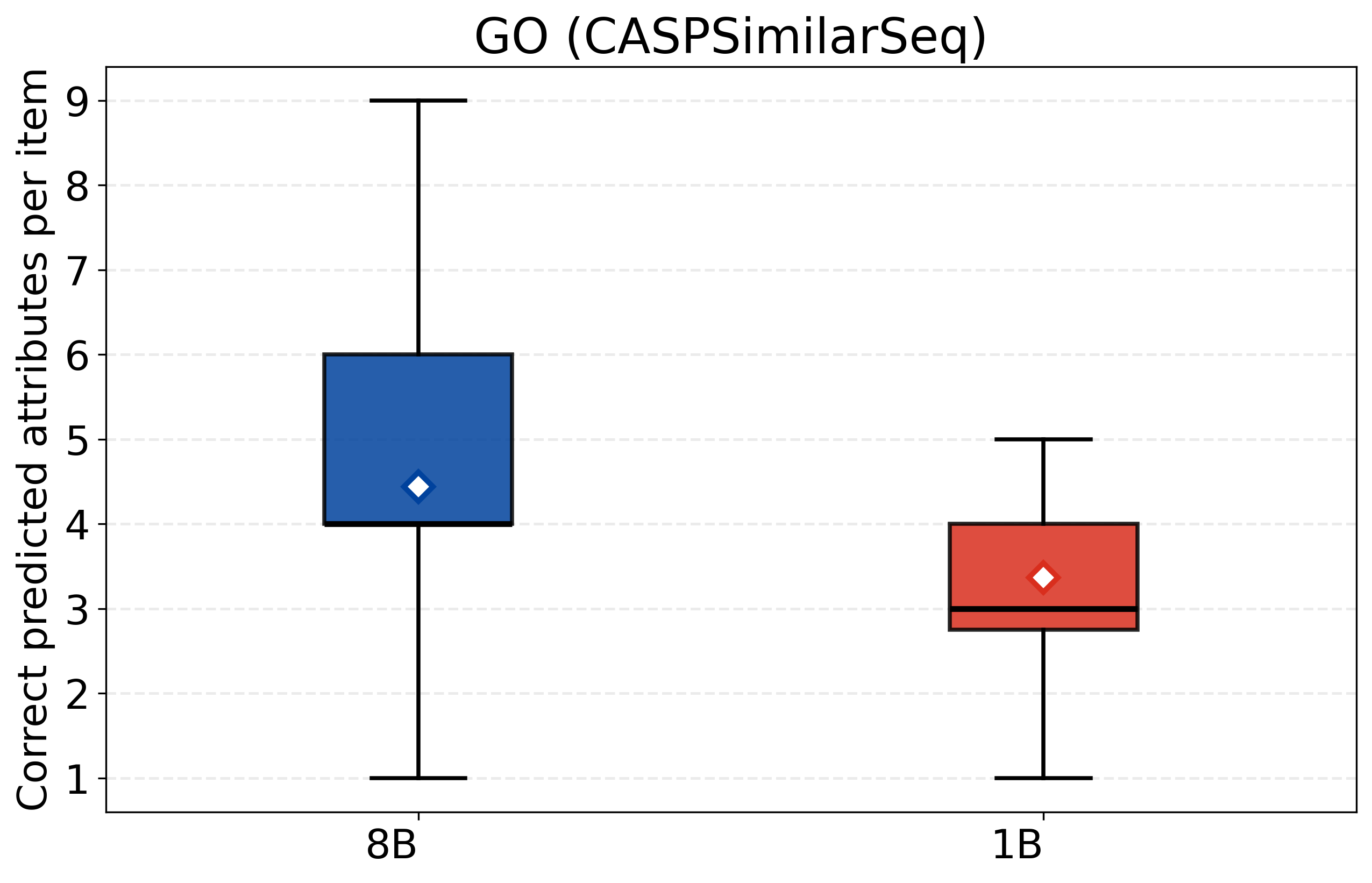
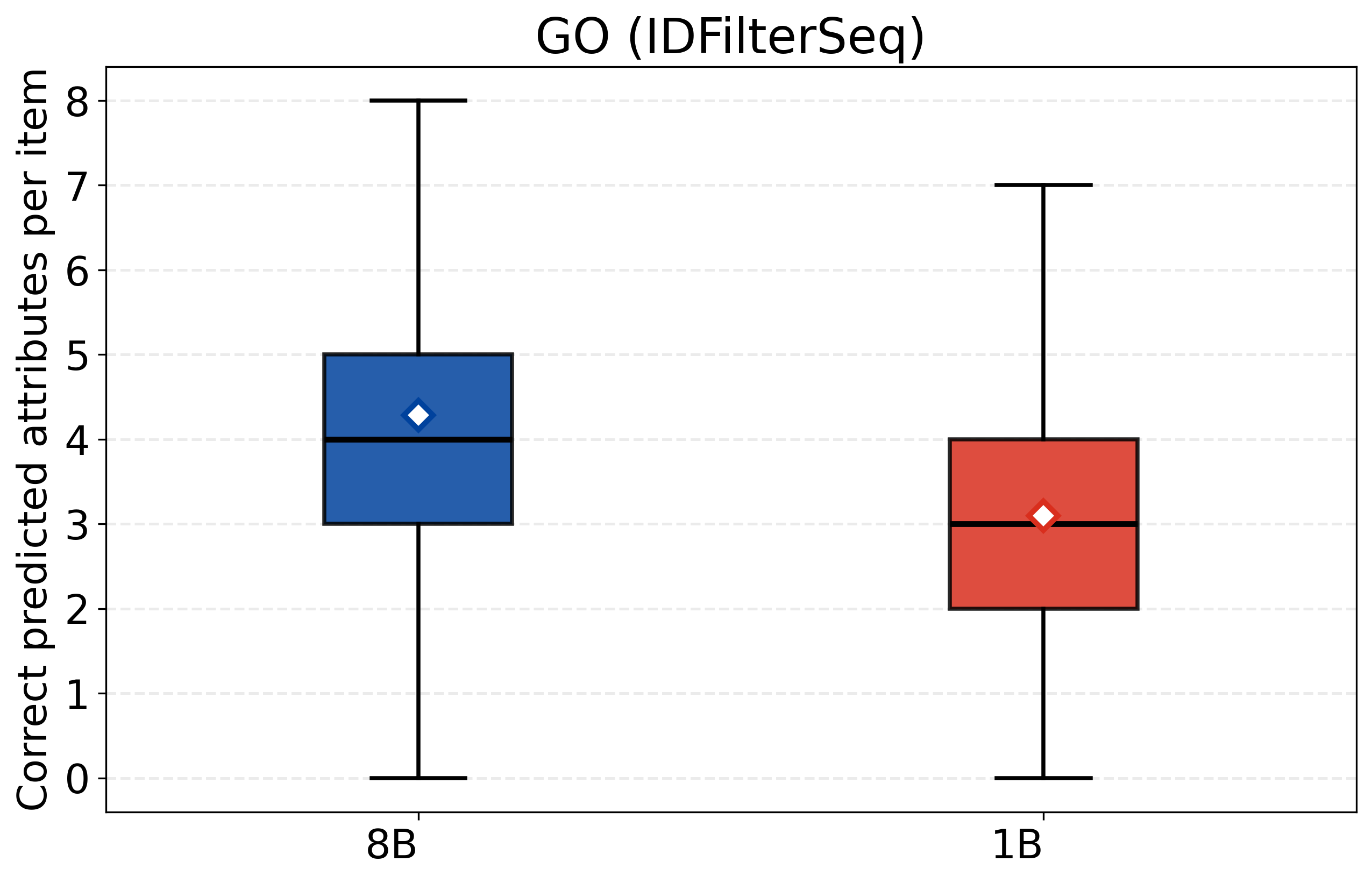
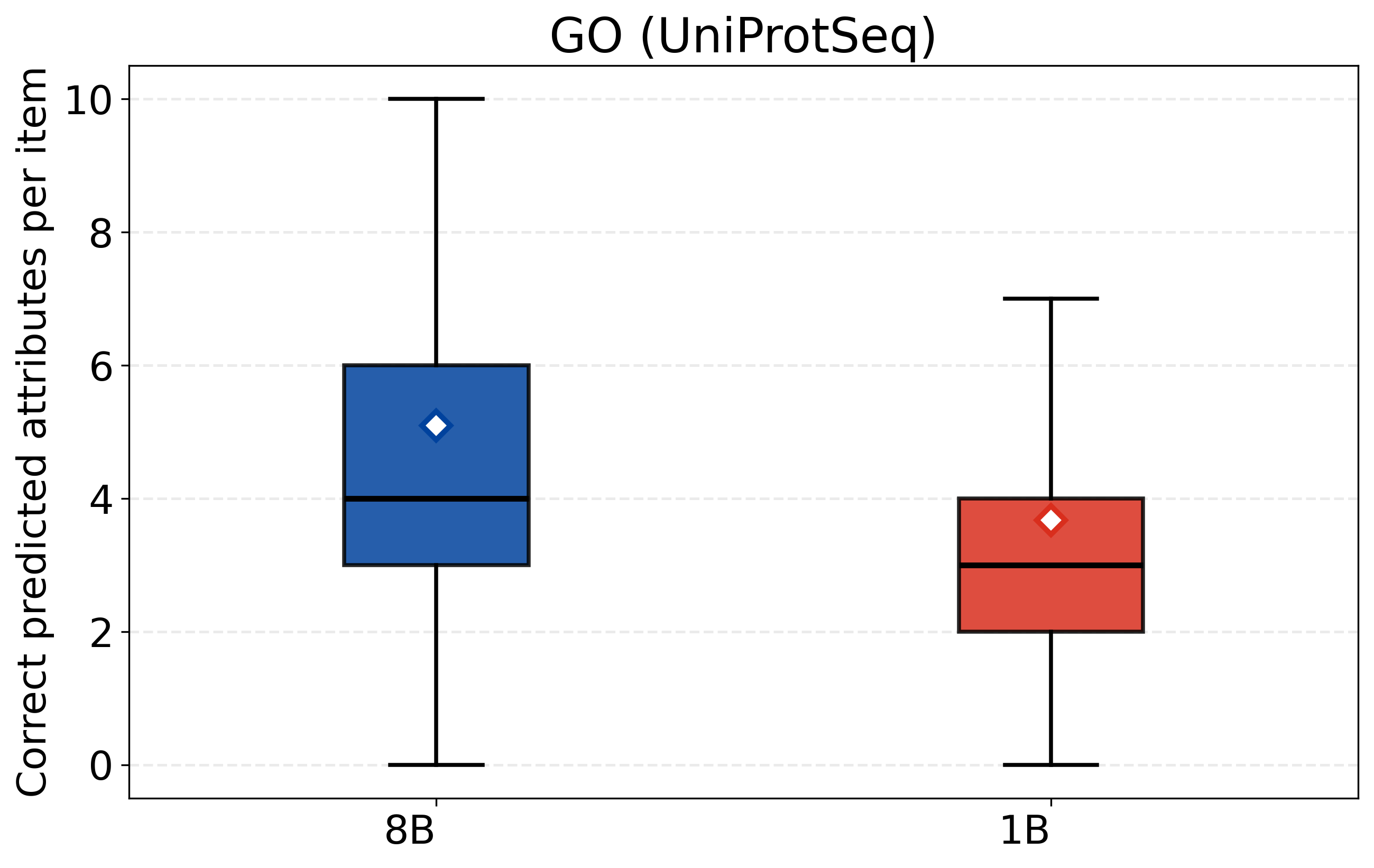
- CASPSimilarSeq: A task assessing protein sequence similarity, aligned with CASP-style evaluations of structure/function. "CASPSimilarSeq"
- Chain-of-thought (CoT): Explicit, step-by-step reasoning traces generated by the model to make its reasoning process transparent. "long-form chain-of-thought"
- Crystallographic Information File (CIF): A standardized text format for storing crystallographic data such as atomic positions and symmetry. "crystal structure files (Crystallographic Information File, CIFs)"
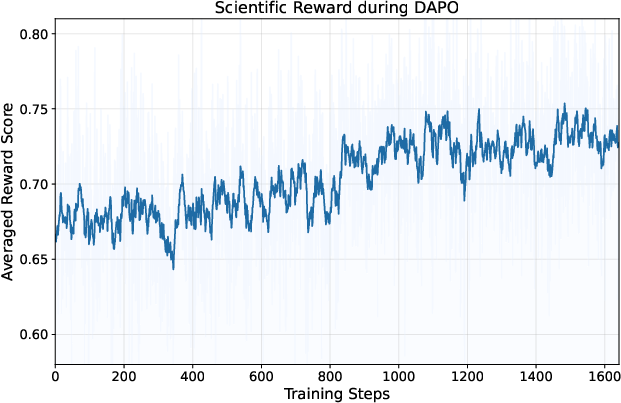
- DAPO: A reinforcement learning algorithm for LLMs that extends PPO with group-based advantages and dynamic sampling. "We adopt DAPO~\cite{yu2025dapo} as the RL algorithm"
- DeepSpeed ZeRO Stage 2: A memory-optimization and parallelization technique enabling large-batch training by partitioning optimizer states and gradients. "DeepSpeed ZeRO Stage 2~\cite{rasley2020deepspeed}"
- Elastic moduli: Quantitative measures of a material’s stiffness (e.g., Young’s modulus), relevant to mechanical behavior. "electronic and mechanical properties (band gap, density, elastic moduli)"
- Empirical solve-rate filter: A data selection method that keeps medium-difficulty items based on observed success rates to improve RL stability. "using an empirical solve-rate filter that targets medium-difficulty instances."
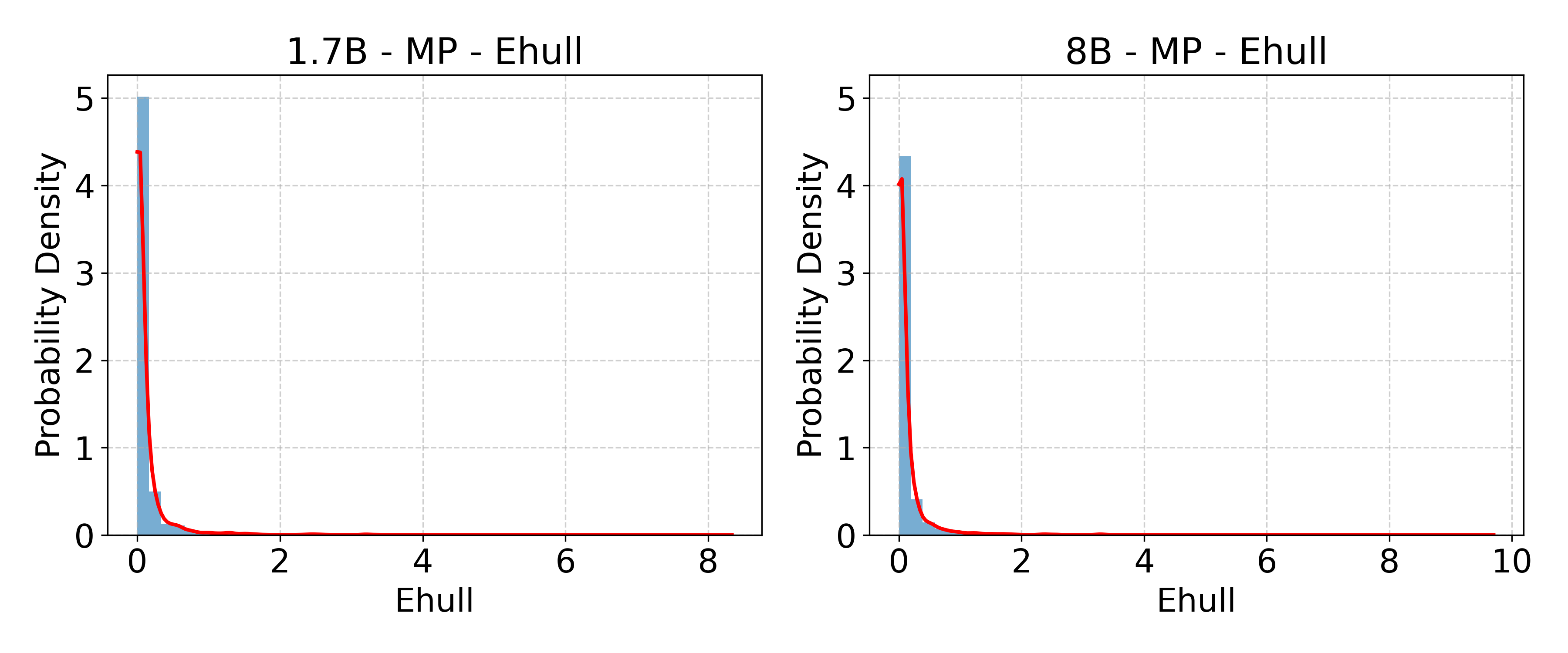
- Energy above hull: A thermodynamic metric indicating a material’s metastability relative to the convex hull of formation energies. "energy above hull"
- FASTA: A text-based format for representing DNA, RNA, or protein sequences with headers and sequence lines. "is represented by the FASTA format."
- FlashAttention: A memory- and speed-optimized attention kernel for transformers reducing I/O overhead. "FlashAttention implementation~\cite{dao2023flashattention}"
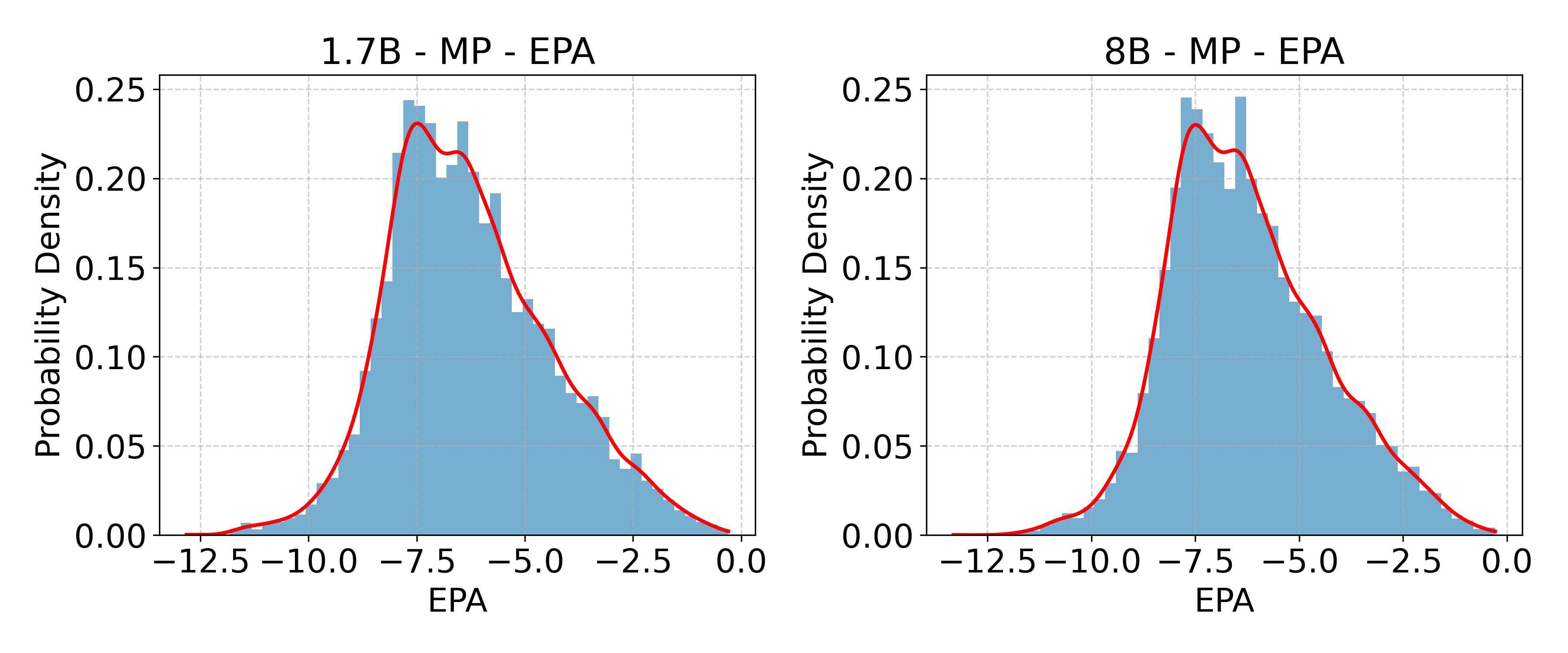
- Formation energy: The energy difference between a compound and its constituent elements, indicating thermodynamic stability. "formation energy"
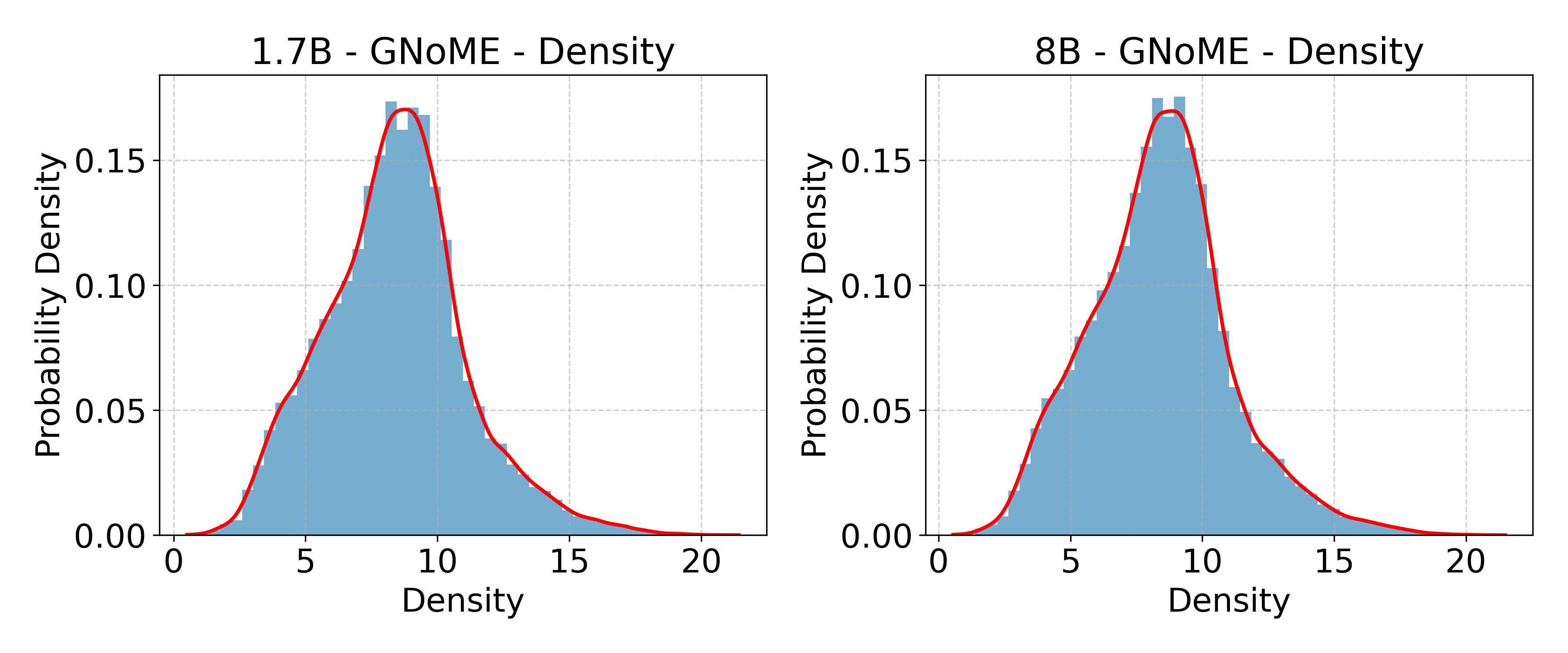
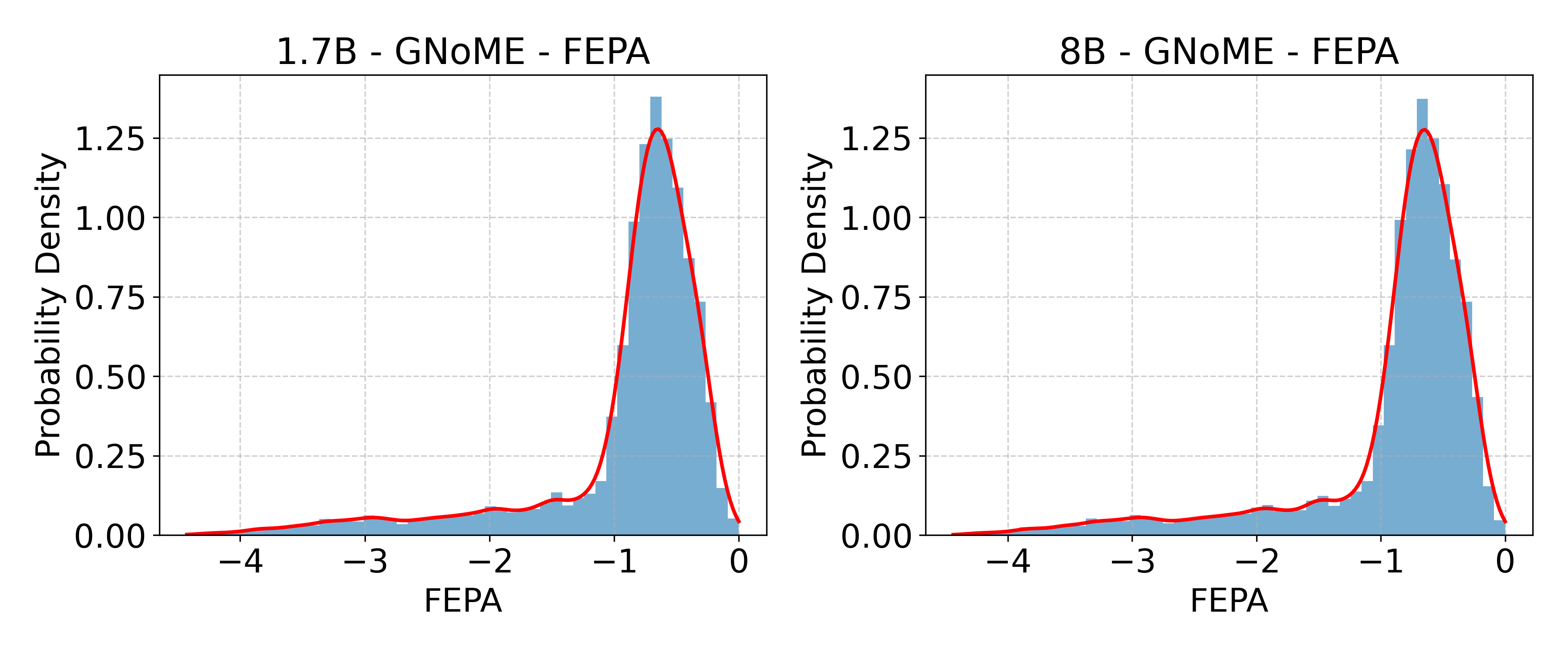
- GNoME: A large-scale materials discovery dataset/system used for training and evaluation in materials science. "GNoME~\cite{merchant2023scaling}"
- Group-standardized advantages: Normalized advantage values computed per candidate group to stabilize policy gradients in RL. "compute group-standardized advantages:"
- GUE: A genomics-related dataset or benchmark used for DNA/RNA modeling and evaluation. "GUE~\cite{zhou2023dnabert}"
- IUPAC: The international authority defining chemical nomenclature; used for standardized molecule names. "International Union of Pure and Applied Chemistry (IUPAC)"
- JARVIS-DFT: A database of density functional theory computed materials properties used for benchmarking prediction tasks. "properties derived from JARVIS-DFT"
- Liger-kernel: Optimized transformer kernels improving training efficiency and throughput. "the Liger-kernel~\cite{hsu2024liger} is enabled for efficiency."
- Materials Project: A public database of computed materials structures and properties for research and ML training. "Material Projects~\cite{materials_project}"
- MENTOR: An evaluation metric for molecular captioning assessing quality and alignment with molecular semantics. "MENTOR"
- Mid-difficulty filter: A selection rule that retains partially solvable examples to avoid trivial or impossible cases during RL. "Mid-difficulty filter. Retain examples with partial success:"
- NCBI: A major repository for biological data including genomes and sequences. "NCBI~\cite{ncbi_homepage}"
- OIG: A large-scale open instruction dataset used for general instruction tuning. "OIG~\cite{oig_laion}"
- OPI: A protein-related dataset used for supervised training/evaluation of biomolecular tasks. "OPI~\cite{xiao2024opi}"
- PEER: A dataset for protein or biology tasks leveraged in supervised fine-tuning. "PEER~\cite{xu2022peer}"
- PPO-style clipped objective: A reinforcement learning objective that clips policy ratios to stabilize updates. "and maximize the PPO-style clipped objective:"
- PubChem: A large chemical database containing molecular structures, properties, and annotations. "PubChem"
- PubMed: A comprehensive database of biomedical literature used for scientific text pretraining. "PubMed~\cite{pubmed}"
- RAbD: Rational Antibody Design; a computational design framework for engineering antibodies. "rational antibody design (RAbD)"
- RDKit: An open-source cheminformatics toolkit used for molecule validation and processing. "RDKit~\cite{rdkit}"
- Reward shaping: Modifying reward signals to guide reinforcement learning toward desired behaviors. "reinforcement learning with task-specific reward shaping"
- Reward softening: Mapping binary rewards into a continuous scale to improve RL convergence and stability. "We therefore replace the binary signal with a reward softening scheme"
- RNAcentral: A database of RNA sequences and annotations used for transcriptomic data collection. "RNAcentral~\cite{rnacentral_homepage}"
- RMSE: Root Mean Squared Error; a regression metric used as a continuous reward signal. "quantitative measures (e.g., RMSE)"
- SELFIES: A robust molecular string representation guaranteed to decode to valid molecules. "Self-Referencing Embedded Strings (SELFIES)"
- SMILES: A line notation encoding molecular graphs as strings for computational processing. "Simplified Molecular Input Line Entry System (SMILES)"
- Solve–check protocol: A procedural workflow that generates solutions and verifies them with retrieval and domain tools. "a solveâcheck protocol with retrieval and domain tools"
- Space-group: A crystallographic symmetry classification describing repeated patterns in crystal structures. "space-group and crystal-system information"
- Stoichiometry consistency: Adherence to correct elemental ratios and balances in chemical predictions or reactions. "stoichiometry consistency"
- Stochastic rollouts: Multiple sampled model outputs used to estimate solve rates and diversity during RL data curation. "Stochastic rollouts."
- Supervised fine-tuning (SFT): Post-training using labeled instructions to align model outputs with task-specific formats. "In the supervised fine-tuning (SFT) stage"
- Task-aware tokenization: A tokenization approach tailored to scientific sequences and representations for unified modeling. "task-aware tokenization"
- Token-level policy-gradient reduction: An RL optimization technique reducing variance by aggregating gradients at the token level. "and a token-level policy-gradient reduction."
- UniRef50: A clustered set of UniProt sequences at 50% identity used to reduce redundancy in protein data. "UniRef50 and UniRef90 (mammalia)"
- UniRef90: A clustered set of UniProt sequences at 90% identity for protein sequence datasets. "UniRef50 and UniRef90 (mammalia)"
- USPTO50k: A dataset of chemical reactions extracted from patents used for synthesis/retrosynthesis tasks. "USPTO50k~\cite{schneider2016s}"
- Valence: The typical bonding capacity of an atom, constraining molecular structures and reactions. "valence"
Collections
Sign up for free to add this paper to one or more collections.