- The paper introduces PCGM to integrate causal relationships and anatomical priors, enhancing the realism of synthetic brain MRIs.
- It leverages a Counterfactual Mask Generator and a 3D ControlNet-based diffusion approach to accurately model subtle neuroanatomical variations.
- Experimental results demonstrate superior image quality, effective age trajectory modeling, and higher anatomical plausibility compared to baseline methods.
Integrating Anatomical Priors into a Causal Diffusion Model
The paper "Integrating Anatomical Priors into a Causal Diffusion Model" presents a novel approach for generating anatomically accurate counterfactual brain MRIs by leveraging causal relationships among brain regions and metadata within a diffusion framework. The objective is to address the limitations of existing counterfactual models that struggle to maintain anatomical plausibility, particularly when generating high-fidelity structural brain MRIs necessary for neurodevelopmental and disease progression studies.
Introduction
Generating anatomically correct brain MRIs is crucial for understanding subtle morphometric differences between subject cohorts, which can be challenging due to high acquisition costs and dataset limitations. The approach proposed, called Probabilistic Causal Graph Model (PCGM), integrates anatomical constraints directly into the generative model to preserve subtle yet medically significant local variations across subjects. PCGM is particularly designed to optimize the synthesis of counterfactual MRIs by incorporating known causal relationships between metadata (e.g., age, sex, disease label) and brain regional measurements.
Probabilistic Causal Graph Model (PCGM) Framework
PCGM features several innovative components, including:
- Probabilistic Graph Module (PGM): Encodes causal relationships using probabilistic graphical models. It models the volume interaction of brain regions based on modified metadata, allowing the simulation of hypothetical anatomical changes due to interventions.
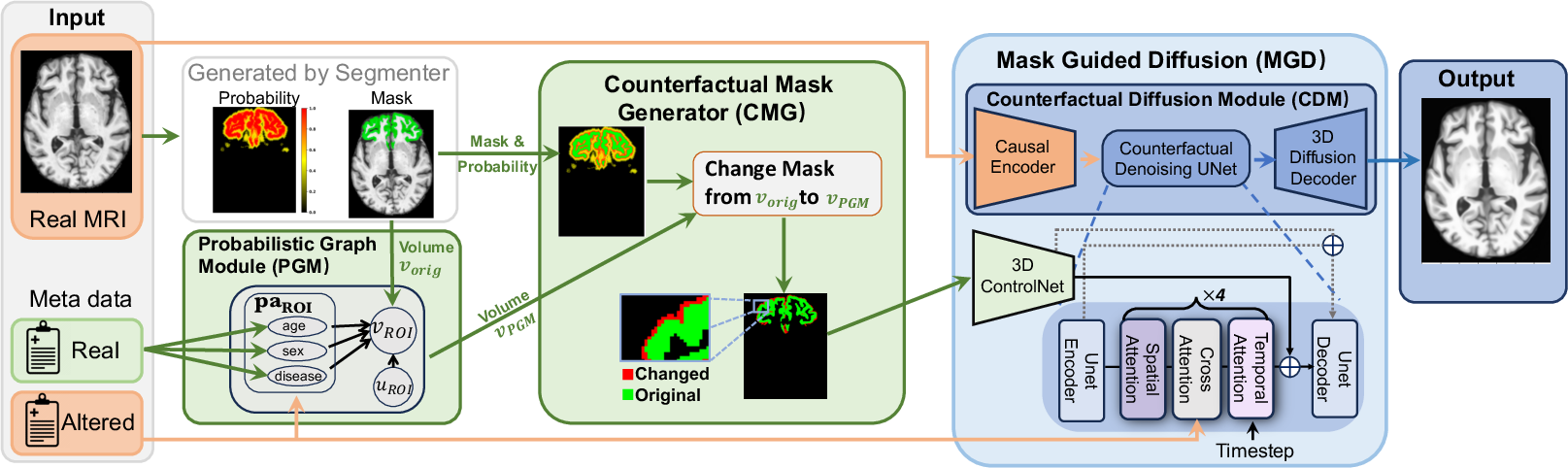
Figure 1: The framework of our Probabilistic Causal Graph Model (PCGM). Known causal relationships between metadata (e.g., age, sex, disease label) and the volumes of brain regions of interest (ROIs) are encoded via the Probabilistic Graph Module (PGM). Given the altered volume of an ROI based on modified metadata (intervention), the Counterfactual Mask Generator\ (CMG) modifies the mask of the ROI extracted from the original MRI to match the new volume score. Based on the original MRI, the metadata, and the counterfactual mask encoded by 3D ControlNet, the Mask Guided Diffusion\ (MGD) module generates the corresponding MRI counterfactuals using our proposed Counterfactual Diffusion Module\ (CDM).
- Counterfactual Mask Generator (CMG): Alters masks based on updated ROI volumes, ensuring changes adhere to anatomical constraints. This is crucial for reflecting subtle variations in regions affected by neuropsychiatric conditions.
- Mask Guided Diffusion (MGD): Generates high-resolution MRIs guided by counterfactual masks and metadata via a 3D extension of ControlNet. It enhances sample fidelity and anatomical accuracy, offering significant improvements over standard 2D operations used in previous models.
Experimental Validation
Comprehensive experiments on multiple datasets demonstrated that PCGM effectively synthesizes structural MRIs with superior quality compared to baseline methods.
- Image Quality: The MRIs generated by PCGM surpassed those synthesized by state-of-the-art GANs and other diffusion models, offering improved image contrast and anatomical detail, especially evident in complicated structures like the cerebellum.
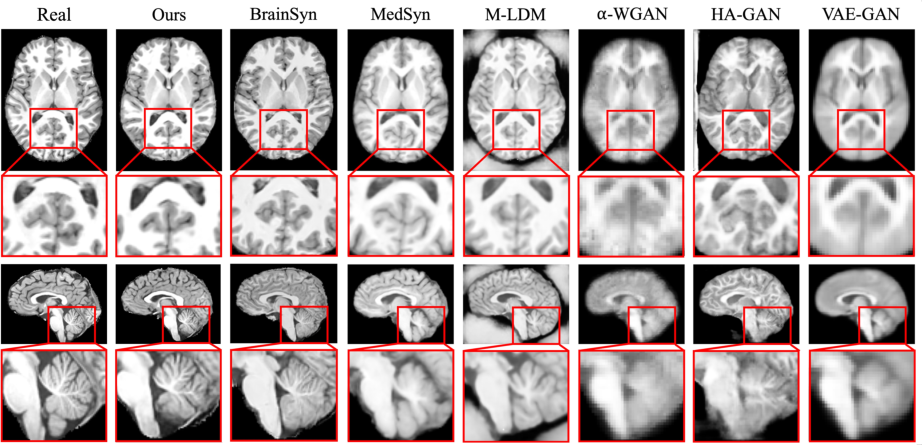
Figure 2: Real and synthetic MRIs created by generative approaches. Given the real MRI, the figure displays the most similar synthetic MRI generated by each method. Each MRI is displayed from the axial (first row) and sagittal view (third row). Compared to baseline approaches, the image contrast and visibility of anatomical structures in the MRI generated by our method most closely match the real MRI, especially for the cerebellum (fourth row).
- Age Trajectory Modeling: PCGM adeptly models the aging trajectory, such as ventricular enlargement, with accuracy far surpassing its competitors, reinforcing its efficacy in generating realistic age-related anatomical changes.
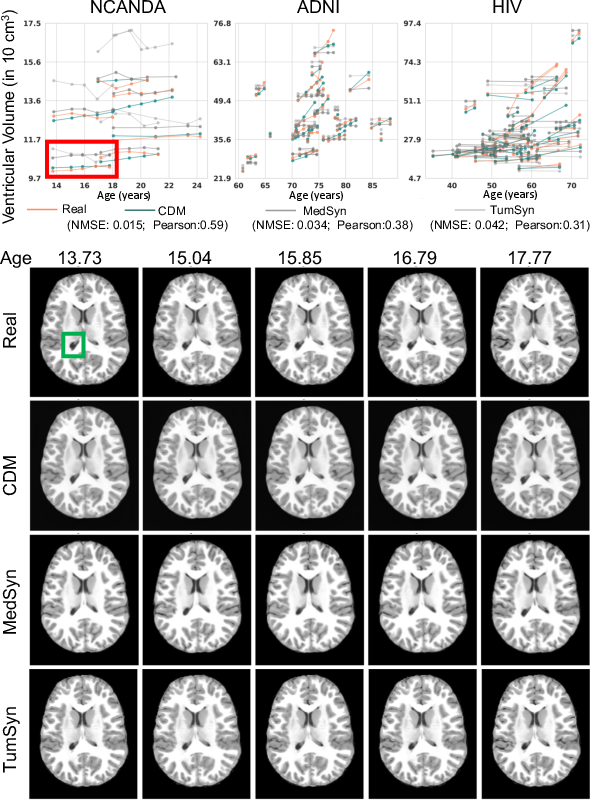
Figure 3: Age trajectory modeling. Each trajectory in the plot represents the volume of the ventricle of one subject measured at different ages. The subjects were participants of NCANDA (ages 14 - 26 years), ADNI (ages 60 - 90 years), or diagnosed with HIV (ages 33 - 74 years). Among the three methods, the trajectories extracted from the counterfactual MRIs produced by our proposed CDM ~align the best (i.e., lowest NMSE and highest Peatson score) with those of the real MRIs. This is also confirmed when visually comparing the longitudinal MRIs of the trajectories outlined in red. Visually the largest differences are shown in the ventricle region outlined in green.
- Anatomical Plausibility: The generated MRIs demonstrated high anatomical plausibility across various brain regions as reflected by the minimal deviations observed in comparative statistical analysis.
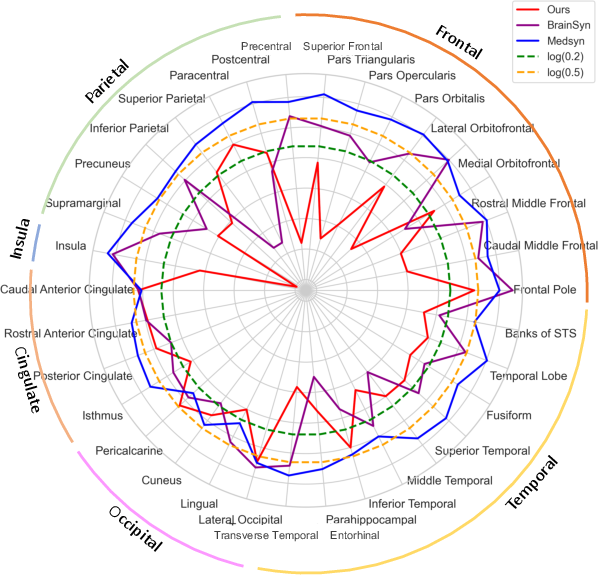
Figure 4: Anatomical Plausibility. Displayed are the natural logarithm of the absolute Cohen's d (|d|) for 32 regions and three methods. |d| \textless 0.2 is viewed as a small effect, 0.2 textless |d| \textless 0.5 indicates a medium effect, and |d|> 0.5 corresponds to a large effect. Our method records the smallest d-score for the majority of regions, indicating the highest overall anatomical plausibility among the 3 methods. Note, the plot connects the scores between ROIs simply to ease comparison.
Conclusion
PCGM represents a significant advancement in generating anatomically accurate counterfactual MRIs, offering unparalleled fidelity and realism crucial for neuroscience research, particularly in studies involving subtle structural changes. By effectively integrating anatomical priors into the generative model, PCGM allows for accurate disease modeling even with limited training data, setting a new benchmark for the synthesis of high-fidelity MRIs in medical imaging applications. The approach not only enhances the potential for synthetic data in augmenting limited MRI datasets but also opens avenues for further research into the application of causal models in medical image analysis. Future work may explore extending these principles to other medical imaging modalities and further refining the causal inference framework to incorporate more complex anatomical and pathological interactions.



