- The paper introduces LLaDA-MedV, a diffusion-based VLM that outperforms traditional ARMs in closed-form biomedical VQA tasks.
- It employs an iterative forward-reverse masked generation process, enabling explicit control over response length and detail.
- Robust training via semantic alignment and vision instruction tuning highlights its potential for generating detailed, visually grounded responses.
LLaDA-MedV: Exploring Large Language Diffusion Models for Biomedical Image Understanding
Introduction
The paper introduces LLaDA-MedV, a diffusion-based vision-LLM (VLM) designed for biomedical image understanding. Autoregressive models (ARMs) have been dominant in this domain, but masked diffusion models (MDMs) like LLaDA-MedV present a promising alternative. LLaDA-MedV achieves consistent performance improvements over existing models such as LLaVA-Med and sets new accuracy benchmarks in closed-form Visual Question Answering (VQA) tasks across several medical datasets.
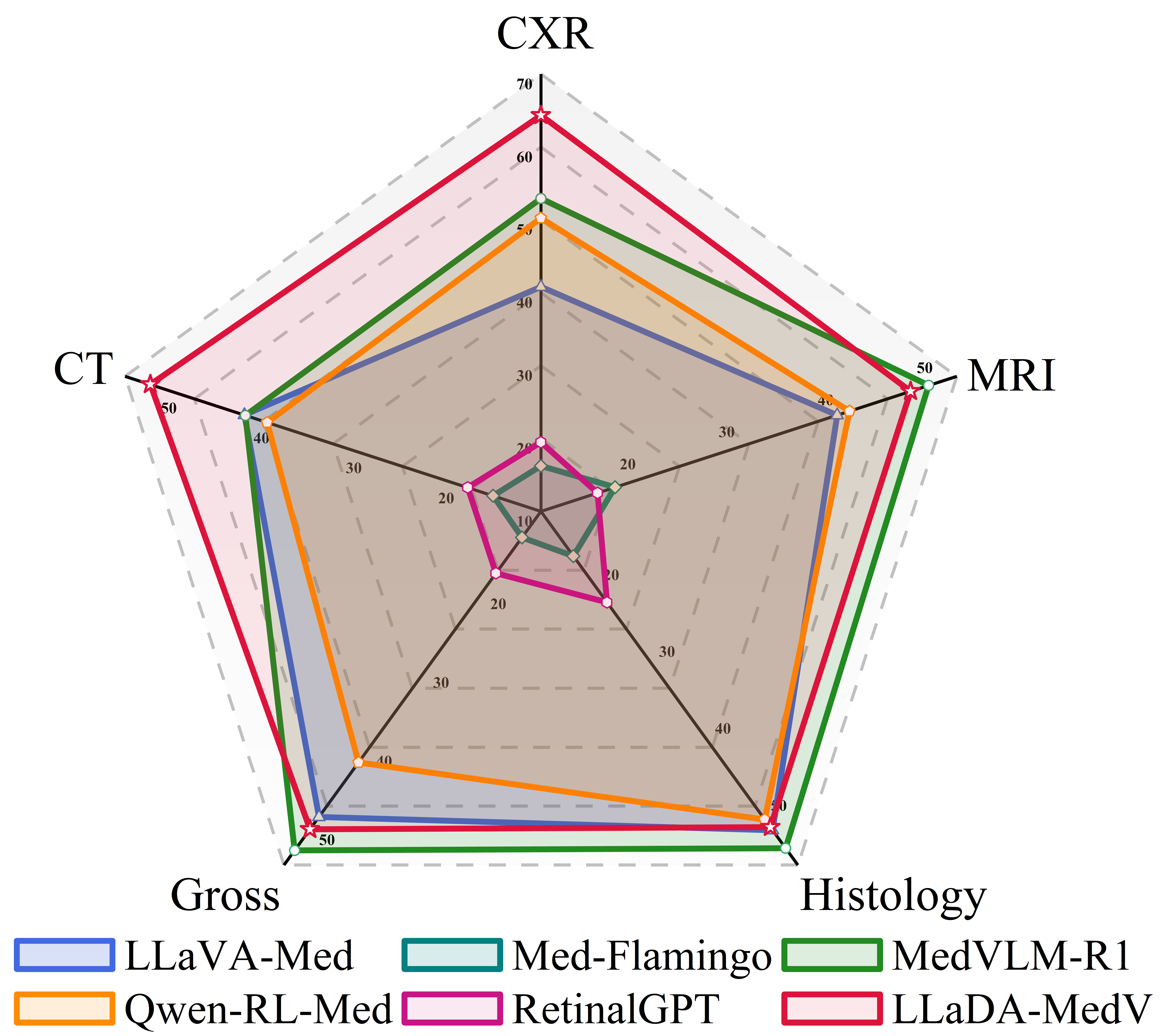
Figure 1: Illustration of biomedical VLMs evaluated in the open-ended biomedical conversation benchmark. Among the 6 Medical VLMs, LLaDA-MedV achieves the highest overall score and demonstrates the best performance on Chest X-ray (CXR) and CT modalities.
Methods
LLaDA-MedV leverages masked diffusion models to operate over discrete tokens. It uses an iterative forward-reverse process where input tokens are masked progressively and then predicted in the reverse generation stage. The approach allows explicit control over response length, enabling longer and more detailed outputs compared to ARMs.
The training involves multiple stages:
- Semantic Alignment: Fine-tuning the projector module to align biomedical language with visual content.
- Vision Instruction Tuning: End-to-end fine-tuning for generating coherent, visually grounded responses.
- Dataset-Specific Fine-Tuning: Enhancement via three biomedical VQA datasets for improved precision.
Experiments and Results
LLaDA-MedV demonstrates superior performance in open-ended biomedical conversations, generating more informative responses with explicit control over output length compared to ARM counterparts such as LLaVA-Med.
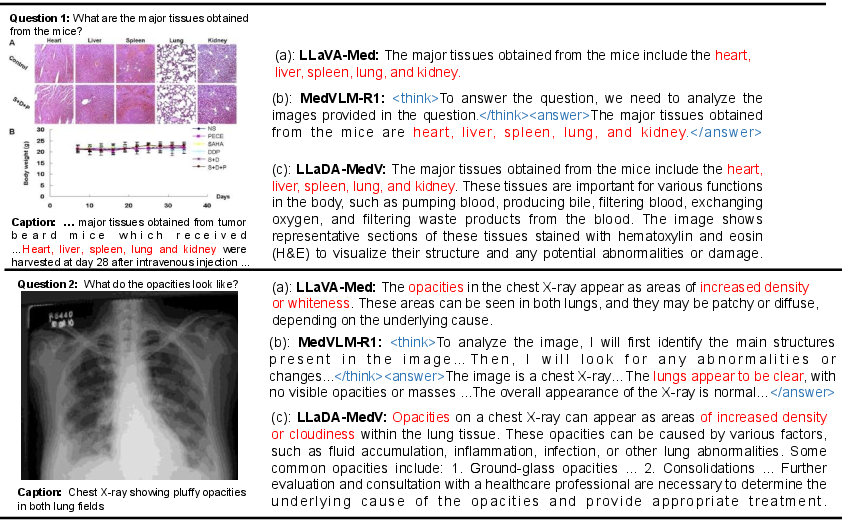
Figure 2: Illustration of open-end conversation evaluation. All questions, images and corresponding captions are sourced from~\cite{li2023llava.
Notably, in downstream VQA tasks, LLaDA-MedV achieves highest accuracy on closed-form queries but faces challenges with open-form questions due to less optimized post-training. This suggests masked diffusion models offer substantial benefits, particularly in controlled response generation scenarios requiring detailed analysis.
Analysis of Training and Inference
The paper reveals critical training factors including proper initialization and fine-tuning strategies as pivotal for performance enhancement. The selection of domain-specific weights significantly impacts model outputs and token repetition issues during inference.

Figure 3: Illustration of LLaVA-Med and LLaDA-MedV responses to biomedical queries 1 and 2. The images, queries, and corresponding captions are adapted from~\cite{li2023llava.
During inference, the trade-off between computational efficiency and response quality is highlighted, with sampling steps being crucial for maintaining response richness while managing computational costs.
Future Work
Token repetition presents a key limitation, especially when the desired output length is large. Future work should focus on optimizing sampling strategies and remasking schedules to balance efficiency and quality, particularly for applications needing detailed responses.

Figure 4: Illustration of token repetition during generation (i.e., marked by red) across different settings. Question 4 and 5 represent the answer from LLaDA-MedV${V_1}.
Conclusion
LLaDA-MedV represents an innovative application of diffusion models for biomedical image understanding. Through strategic training and inference methodologies, it presents a compelling alternative to traditional ARMs, promising improved response control and quality in biomedical AI applications. Further research should address the optimization challenges to fully leverage diffusion models in the field of medical image analysis.



