- The paper presents an open-source framework that curates over 24M biomedical image-caption pairs from PubMed Central, enabling efficient VLM training.
- It employs a sophisticated curation pipeline that extracts metadata and clusters images into 12 global and 170 local concepts with 27 unique metadata fields.
- The BMCA-CLIP models trained on BIOMEDICA achieve up to 29.8% improvement in dermatology tasks while reducing compute requirements by 10x.
BIOMEDICA: An Open Biomedical Image-Caption Archive, Dataset, and Vision-LLMs Derived from Scientific Literature
Introduction
The development of generalist vision-LLMs (VLMs) for biomedical domains has been impeded by the lack of extensive and publicly accessible datasets that span the diverse fields of biology and medicine. "BIOMEDICA: An Open Biomedical Image-Caption Archive, Dataset, and Vision-LLMs Derived from Scientific Literature" (2501.07171) presents a solution by offering an open-source framework that curates over 24 million unique image-text pairs from the PubMed Central Open Access subset, encompassing more than 6 million articles. By leveraging this extensive dataset, the framework enables the creation of generalized biomedical VLMs that promise expert-level performance across numerous tasks while requiring substantially less computational resource.

Figure 1: Overlap of BIOMEDICA dataset with the landscape of biomedical research, highlighting the diversity of topics covered.
Curation and Scope of BIOMEDICA
BIOMEDICA employs a sophisticated curation pipeline to extract and organize metadata, captions, and images from the massive corpus of the PMC-OA repository. The pipeline involves several key stages: extraction of metadata and initial data processing, feature generation using DINO v2, and annotation of clusters by domain experts, resulting in the identification of 12 global concepts and 170 finer-grained local concepts.
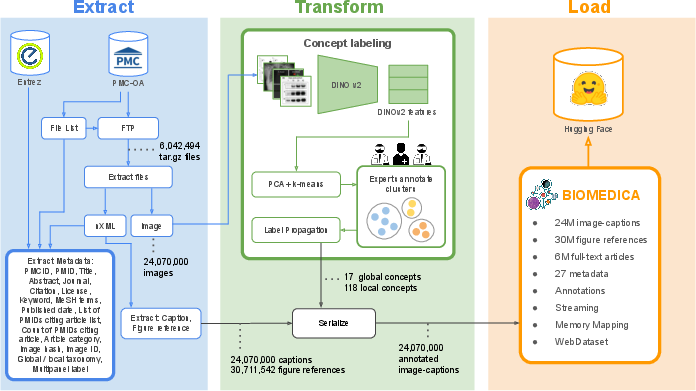
Figure 2: BIOMEDICA curation pipeline showcasing the extract, transform, and load stages.
Each data point in the BIOMEDICA dataset is enriched with over 27 unique metadata fields, facilitating detailed querying and filtering. For computational efficiency, the dataset is stored in a dual format, supporting both high-throughput streaming via WebDataset and querying through Parquet, achieving notable input/output rate improvements.
The release of BMCA-CLIP models underscores the applicability of BIOMEDICA in training VLMs. These models are fine-tuned via streaming on the BIOMEDICA dataset, which avoids the need for local downloading of the voluminous data. The models achieve notable performance gains, with an average improvement of 6.56% over prior state-of-the-art models, reaching up to 29.8% in dermatology tasks alone.
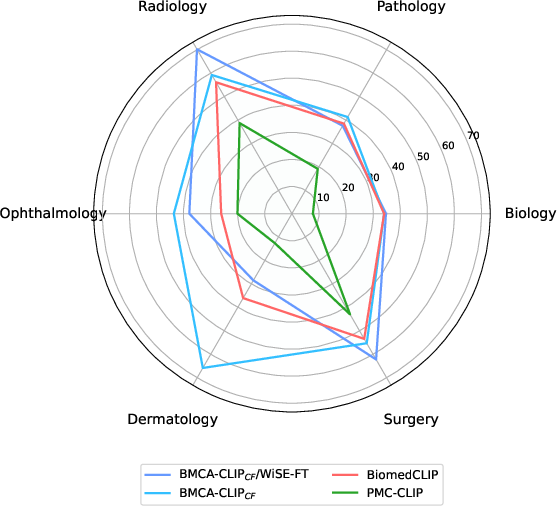
Figure 3: Average model performance of best BMCA-CLIP models compared to prior work, illustrating significant advancements across different biomedical tasks.
This performance is coupled with a reduction in computational demand, utilizing 10 times less compute compared to predecessor models, thanks to the strategic data curation and high data throughput achieved by BIOMEDICA's architecture.
Evaluation and Deployment Implications
The development outcomes include extensive evaluation benchmarks covering 40 established biomedical datasets, which exhibit comprehensive assessment properties across pathology, radiology, molecular biology, and more. This ensures a robust evaluation of the VLMs against a comprehensive suite of retrieval and classification tasks, affirming their effectiveness and generalizability.
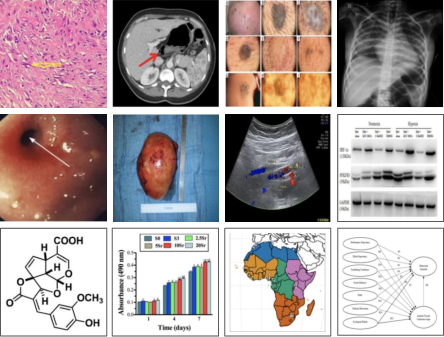

Figure 4: A selection of diverse image types from the BIOMEDICA dataset and the structure of its taxonomy.
The models are optimized for zero-shot inference, crucial for scenarios where computational resources are limited, and where rapid deployment is critical. Therefore, BIOMEDICA offers a significant contribution to the democratization of AI in biomedicine, providing a comprehensive resource for developing highly efficient, adaptable models across a variety of biomedical domains.
Conclusion
BIOMEDICA presents a critical step forward in the domain of biomedical vision-language modeling, offering a unique dataset with an extensive scale, rich metadata, and optimized access strategies, invaluable for both foundational model training and downstream application development. Future endeavors may focus on expanding the dataset and exploring further integration with other biomedical resources to enhance model diversity and accuracy. As the dataset is available to the research community, BIOMEDICA is positioned to significantly advance the field of biomedical AI across both academia and industry fronts.






