- The paper introduces a novel fine-grained vision-language pre-training method that aligns CT image anatomical regions with radiology reports.
- It leverages detailed anatomical segmentation and report decomposition, achieving an average AUC of 81.3% in zero-shot abnormality detection across 54 tasks.
- A dual false negative reduction strategy improves model accuracy by effectively handling misalignments between normal and abnormal anatomical features.
Fine-Grained Vision-Language Pre-training for CT Image Understanding
The paper "Large-scale and Fine-grained Vision-language Pre-training for Enhanced CT Image Understanding" (2501.14548) introduces a fine-grained vision-LLM (fVLM) designed for anatomy-level CT image interpretation. This approach addresses limitations in existing contrastive language-image pre-training (CLIP) methods by explicitly matching anatomical regions in CT images with corresponding descriptions in radiology reports. The paper highlights a novel dual false negative reduction approach to handle challenges arising from the abundance of healthy anatomy samples and similarly diseased abnormalities. The experiments conducted on a newly curated large-scale CT dataset and publicly available benchmarks demonstrate the potential of fVLM for versatile medical image interpretation.
Motivation and Background
The increasing demand for AI in medical image interpretation is driven by the need to assist radiologists in efficiently and accurately diagnosing a wide range of conditions [udare2022radiologist, blankemeier2024merlin]. Supervised learning, while successful in natural scene images, faces challenges in the medical domain due to the complexity and variability of medical conditions, as well as the labor-intensive annotation process [isensee2021nnu, wang2023sammed3d, zhang2023parse, guo2024towards]. Vision LLMs (VLMs) offer a promising alternative by leveraging radiology reports as a natural supervision source, eliminating the need for explicit disease category labels [cao2024bootstrapping]. However, current VLMs typically employ global contrastive learning, which contrasts entire images with reports, neglecting local associations between imaging regions and report sentences [bai2024m3d, hamamci2024foundation]. This coarse-grained approach can lead to misalignment issues and compromise model performance and interpretability.
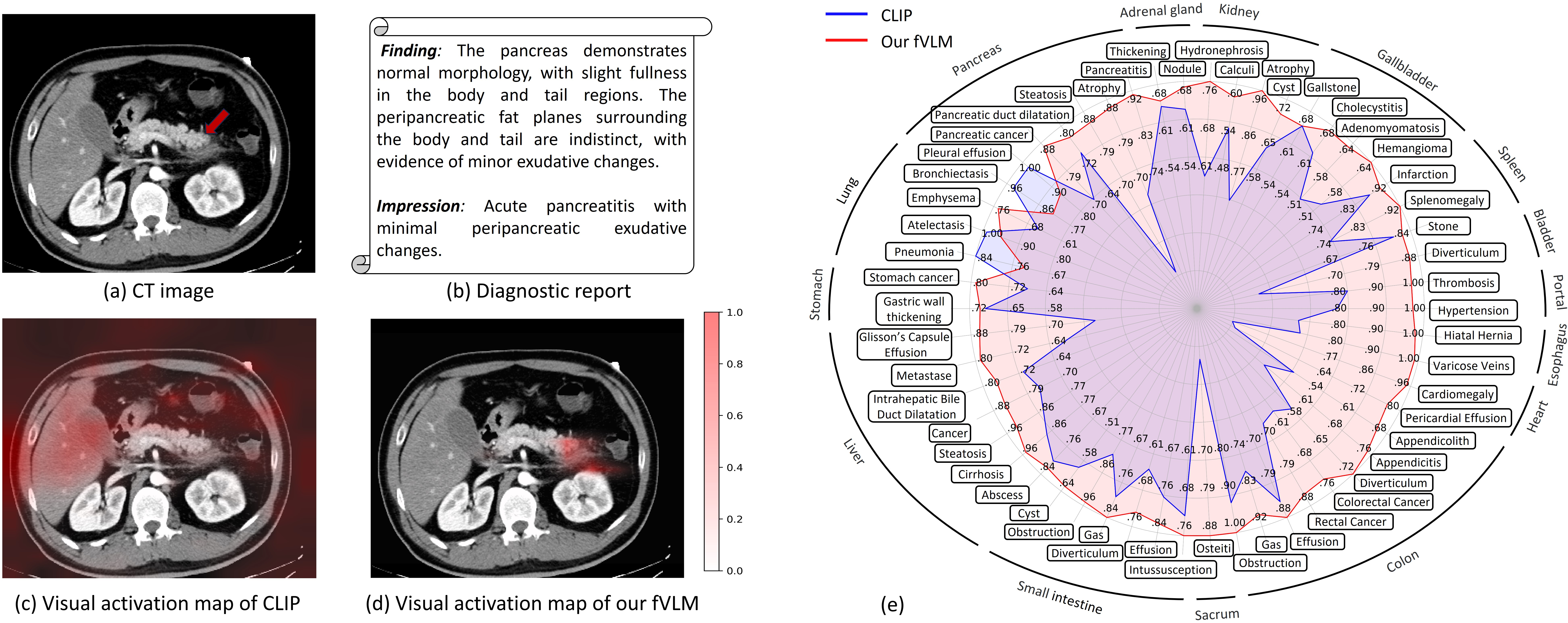
Figure 1: Comparative analysis of vanilla VLM (CLIP) and fVLM, illustrating regions of interest for pancreatitis diagnosis and quantitative AUC score comparisons across 54 disease diagnosis tasks.
Methodology: Fine-Grained VLP
The proposed fVLM moves beyond global image-text contrastive learning by enabling anatomy-level fine-grained alignment between CT scans and reports. The approach involves decomposing both images and reports at the anatomical level and then performing contrastive pre-training for each anatomy individually (Figure 2).
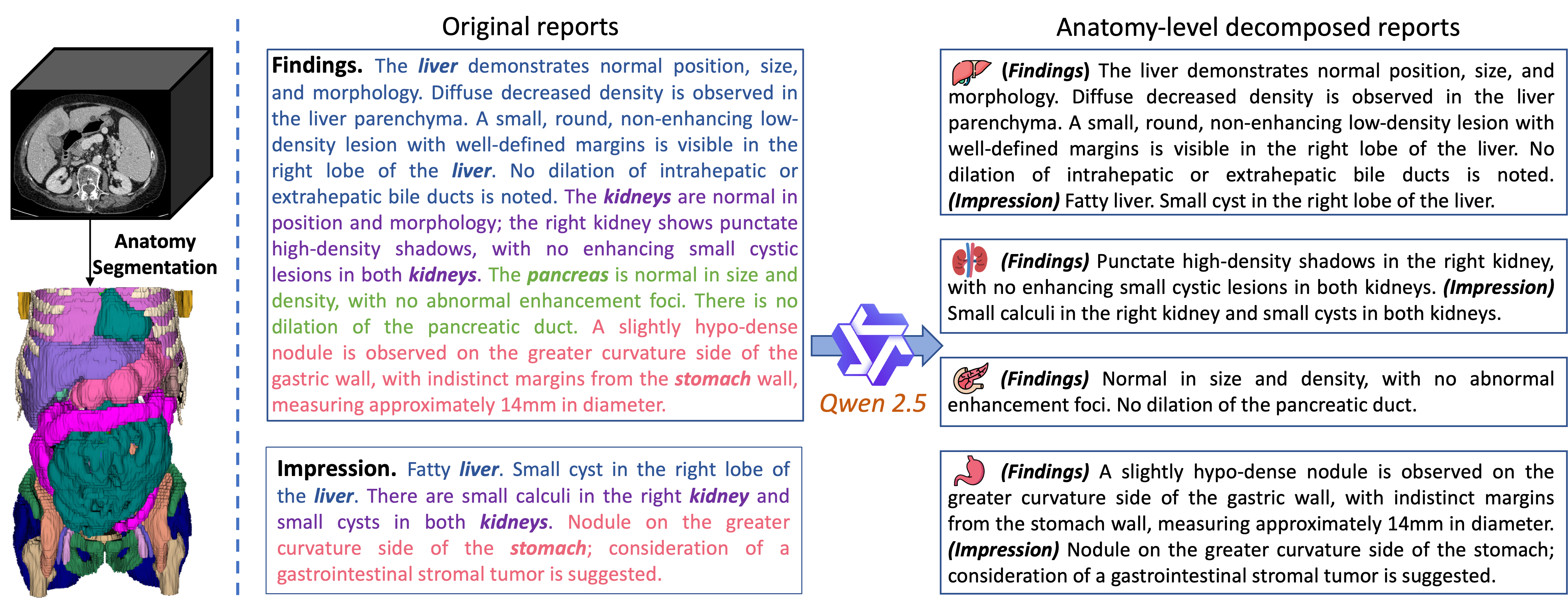
Figure 3: Illustration of CT anatomy parsing and diagnostic report decomposition.
The process includes:
- Anatomy Parsing: Utilizing Totalsegmentator to generate detailed anatomical structure masks for 104 regions within CT scans [wasserthal2023totalsegmentator]. These regions are then grouped into 36 major anatomies to align with the granularity of descriptions in clinical reports.
- Report Decomposition: Decomposing raw CT diagnostic reports according to the grouped anatomies using a divide-and-conquer strategy. An LLM, Qwen 2.5 [qwen], is used to identify anatomies mentioned in the findings and impression sections of each report and extract anatomy-specific descriptions.
- Fine-grained Contrastive Pre-training: Adapting the CLIP architecture [radford2021learning] with vision transformer (ViT) [dosovitskiy2020image] and BERT [devlin2018bert] as image and text encoders, respectively. The image encoder transforms the input CT volume into a compact visual embedding. For each anatomy, a segmentation mask guides the construction of anatomy-specific visual representations. A learnable anatomy-wise query token is appended to the extracted tokens and updated via a self-attention layer. The updated query token generates anatomy-wise visual representation.
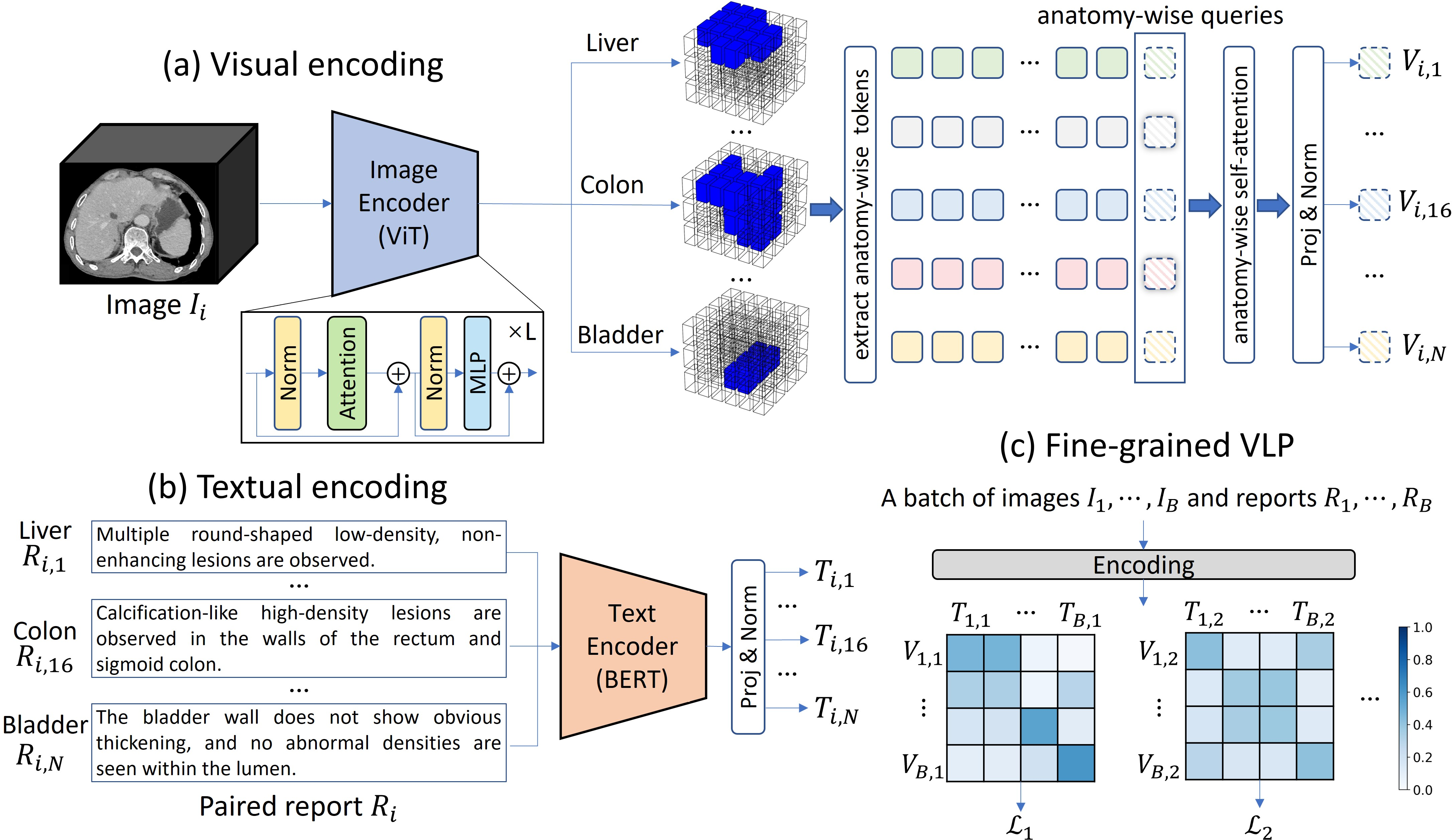
Figure 2: Framework of fVLM, illustrating visual encoding, textual encoding, and fine-grained VLP processes.
Addressing False Negatives
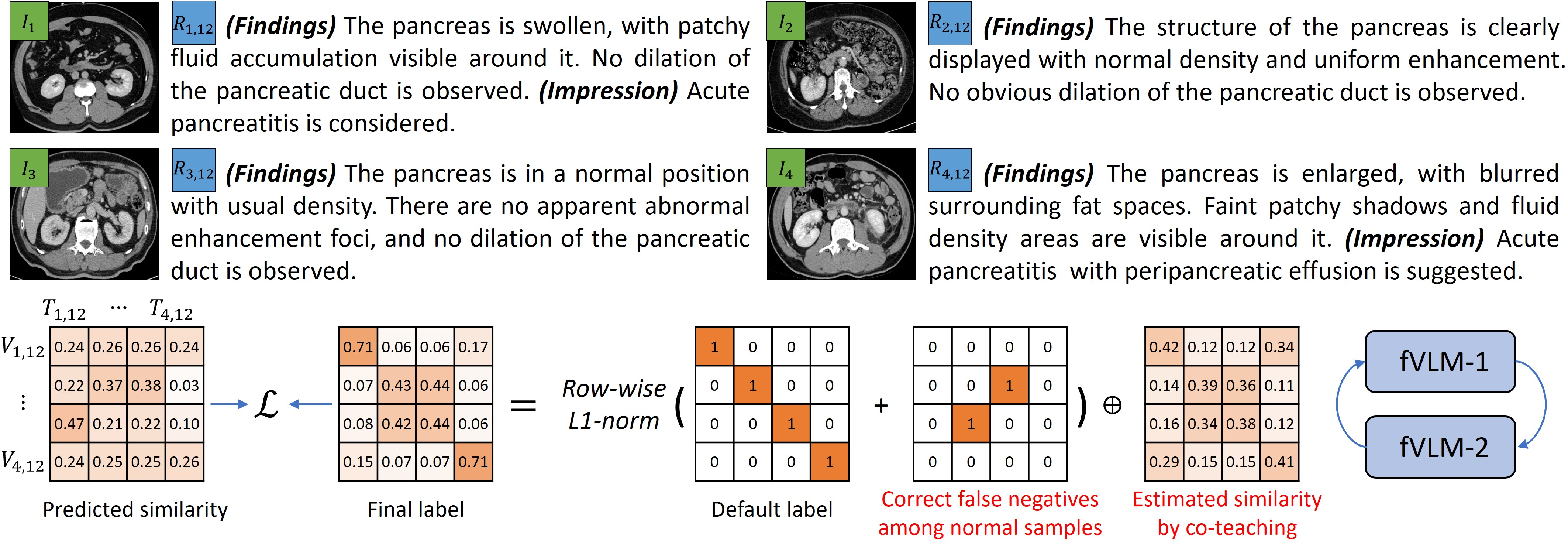
Fine-grained alignment faces challenges related to false negatives, primarily due to the prevalence of healthy anatomy samples and similar abnormalities across different diseases. The paper introduces a dual false negative reduction (FNR) approach to address this issue (Figure 4):
Experimental Results
The fVLM was evaluated on a newly curated large-scale CT dataset, MedVL-CT69K, comprising 272,124 CT scans from 69,086 patients. The model's performance was assessed on zero-shot abnormality detection and downstream report-generation tasks.
- Zero-shot Abnormality Detection: fVLM achieved an average AUC of 81.3\% across 54 disease diagnosis tasks, surpassing CLIP by 12.9\% and a supervised baseline by 8.0%. On the CT-RATE and Rad-ChestCT benchmarks, fVLM outperformed the state-of-the-art approach by 7.4\% and 4.8\% absolute AUC gains, respectively.
- Radiology Report Generation: In downstream report generation tasks, fVLM demonstrated superior performance compared to models with purely visual pre-training, highlighting the benefits of aligning visual and textual features.
The visualization results show that fVLM has the capacity to precisely localize pathological changes across a spectrum of conditions.
Implications and Future Work
The fVLM presents a scalable and annotation-free approach for CT image interpretation, demonstrating strong scaling capabilities and addressing vision-language misalignment issues through fine-grained anatomy-level contrastive learning. The dual false negative reduction module effectively alleviates the adverse effects of false negatives in both normal and abnormal samples. These advancements contribute to improved diagnostic accuracy and interpretability in medical imaging.
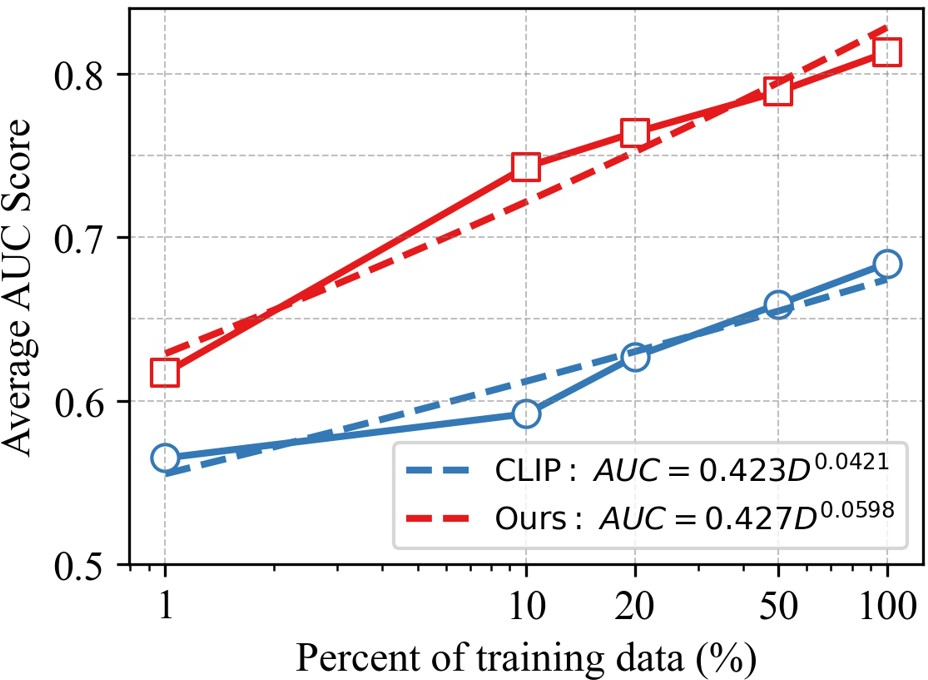
Figure 5: Scaling laws of CLIP and fVLM, showing performance improvements as training data increases.
Future work may explore anatomy-wise report generation to fully leverage the potential of fVLM. Overcoming the resource consumption and time commitment associated with localizing anatomical structures and decomposing diagnostic reports remains a key challenge for implementation.