- The paper introduces SAMed-2, which leverages a temporal adapter and a confidence-driven memory mechanism to overcome limitations in multi-modal medical imaging.
- It integrates spatial-temporal techniques using 3D convolutions and attention mechanisms, significantly enhancing noise robustness and mitigating catastrophic forgetting.
- Experiments on the MedBank-100k dataset across 21 tasks demonstrate superior performance with higher Dice scores and effective zero-shot generalization.
SAMed-2: Selective Memory Enhanced Medical Segment Anything Model
Introduction
The paper "SAMed-2: Selective Memory Enhanced Medical Segment Anything Model" presents a novel approach to adapting general-purpose segmentation models for complex medical imaging tasks. Medical image segmentation is pivotal for diagnostic and therapeutic processes in clinical practice. Traditional models like U-Net demand extensive retraining for each new application due to the diverse nature of medical imaging, which poses significant challenges due to data annotation costs and variance in imaging modalities.
SAMed-2 is designed to overcome these limitations by building upon the SAM-2 architecture. The model integrates a temporal adapter within the image encoder to capture spatial and temporal correlations, and a confidence-driven memory mechanism that enhances robustness against noise and mitigates catastrophic forgetting. This paper also introduces MedBank-100k, a dataset encompassing seven imaging modalities and 21 tasks, to train and benchmark SAMed-2.
Methodology
Architecture Overview
SAMed-2's architecture is rooted in SAM-2, which segments images by embedding input data and managing previous segmentations through a memory base. The enhancements introduced in SAMed-2 aim to specifically address medical image segmentation challenges.
Temporal Adapter
The temporal adapter facilitates the integration of temporal knowledge from medical datasets, which often include volumetric or time-sequential images like CT and MR slices. This adapter uses a combination of spatial attention mechanisms and 3D convolutions to capture multi-dimensional context effectively.
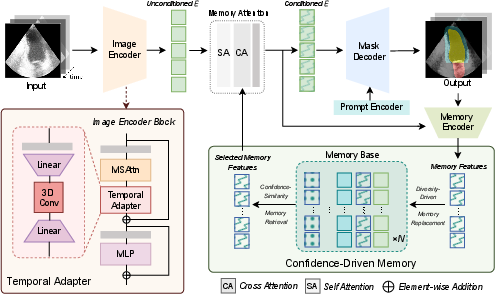
Figure 1: Workflow of SAMed-2. It integrates a temporal adapter in the image encoder to capture multi-dimensional context and a confidence-driven memory module to store high-certainty features.
Confidence-Driven Memory Mechanism
Addressing noise in training datasets is crucial for effective model performance. SAMed-2 introduces a memory module that retains only high-certainty features, enabling the model to generalize across different modalities without the model forgetting learned tasks. The memory retrieval process selects features based on both similarity and confidence, improving noise robustness and overall performance.
Implementation and Dataset
The MedBank-100k dataset is curated to provide a wide variety of medical scenarios, further enhancing the model's ability to handle real-world applications. The data spans fundus, dermoscopy, X-ray, CT, MR, colonoscopy, and echocardiography modalities.
Experimental Results
The effectiveness of SAMed-2 is demonstrated through both internal validations across 21 tasks and external validations on unseen datasets. The experiments confirm that SAMed-2 outperforms existing state-of-the-art models, particularly in multi-task scenarios.


Figure 2: Few-shot scaling result of our method and compared methods on external prostate segmentation task.
Quantitative Performance
The model showcases superior performance with an average Dice Similarity Coefficient (DSC) surpassing other models such as MedSAM-2, SAM-2, and U-Net. Notably, SAMed-2 improves zero-shot performance on external datasets by a significant margin, demonstrating its versatility and robustness.
Ablation Studies
Ablation studies confirm the importance of the confidence-driven memory mechanism and temporal adapter in achieving the model's performance, with each component providing specific advantages in handling various imaging challenges.
Conclusion
SAMed-2 represents a significant advancement in medical image segmentation by effectively adapting the segment-anything paradigm to the complex and varied nature of medical imaging tasks. Its architecture, designed explicitly for handling multi-dimensional and noise-prone datasets, sets a new benchmark in medical image analysis. The introduction of MedBank-100k further strengthens the domain-specific utility of SAMed-2, providing a comprehensive resource for future model training and evaluation.
The research outlines pathways for future exploration, including memory mechanisms' further refinement and exploring other architectures to enhance temporal and spatial learning. This work underscores the potential of foundational segmentation models in advancing clinical practice’s technological capabilities.


