- The paper introduces CBR-DDI, a novel framework integrating case-based reasoning with LLMs and GNNs to enhance accuracy and interpretability in drug-drug interaction predictions.
- It employs a hybrid retrieval method combining semantic and structural similarities to build a knowledge repository from historical drug case data, achieving a 28.7% accuracy improvement on benchmarks.
- The approach provides actionable clinical insights through dual-layer knowledge-enhanced prompting that transparently synthesizes factual drug associations and historical interaction mechanisms.
Enhancing Drug-Drug Interaction Prediction with Case-Based Reasoning
This essay presents a detailed analysis of the framework proposed in the paper "Case-Based Reasoning Enhances the Predictive Power of LLMs in Drug-Drug Interaction" (2505.23034). The research addresses the challenge of predicting drug-drug interactions (DDIs) using a novel method that leverages historical case data within a LLM environment. The method, termed CBR-DDI, integrates Case-Based Reasoning (CBR) with LLMs and Graph Neural Networks (GNNs) to improve predictive performance and interpretability in DDI tasks.
Framework Overview
CBR-DDI is developed to address the limitations of current LLM approaches in DDI prediction, which often lack interpretability and struggle to generalize predictions to novel drugs. By utilizing CBR, CBR-DDI constructs a knowledge repository that encapsulates pharmaceutical principles distilled from historical cases. The integration of LLMs for extracting pharmacological insights and GNNs for modeling drug associations provides a robust framework for knowledge-enhanced reasoning.
The knowledge repository is foundational to this method, comprising structured representations of numerous drug cases. Each case includes functional descriptions generated by LLMs, drug associations encoded through GNNs, historical interaction mechanisms, and interaction labels. This repository enables the hybrid retrieval of semantically and structurally similar cases, which are pivotal during the prediction phase.
Case-Based Reasoning Process
Case Retrieval
The retrieval process employs a hybrid mechanism, integrating LLMs for semantic similarity and GNNs for structural similarity. The retrieval score is computed as a weighted sum of both components, allowing for comprehensive identification of relevant cases. This balance ensures that retrieved cases do not merely reflect data proximity but also encapsulate shared pharmacological patterns.
Knowledge-Enhanced Prompting
In the reasoning phase, retrieved cases serve as inputs to a dual-layer knowledge-enhanced prompt, which the LLM uses to synthesize factual and regularity knowledge. By incorporating both external drug association and historical interaction mechanisms into the prompt, the LLM can produce accurate and interpretable interaction predictions. This process is further refined by filtering candidate interaction types, thus focusing the model's reasoning capabilities.
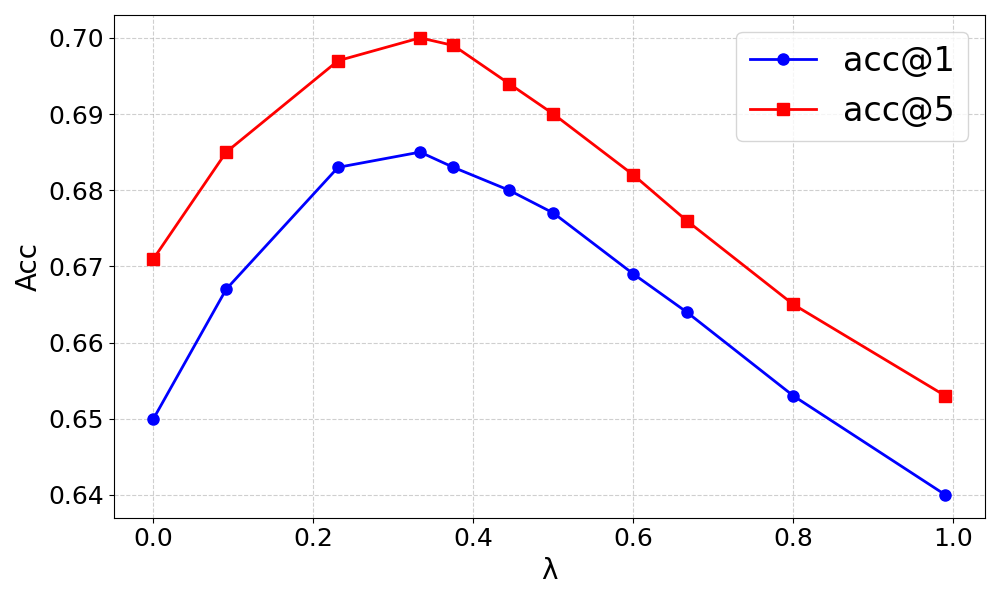
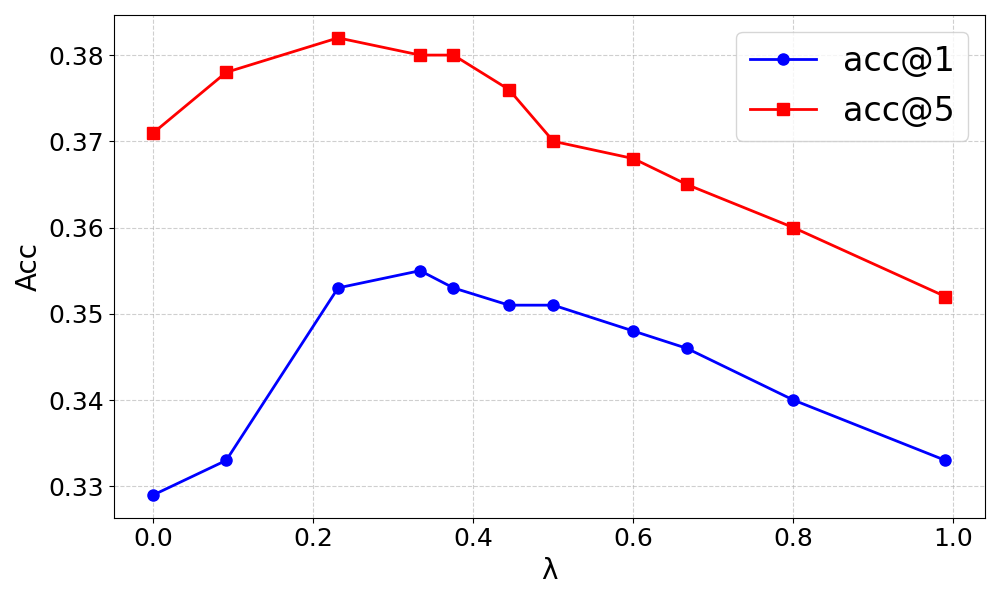
Figure 1: Impact of the hyperparameter λ in the hybrid retriever on accuracy for DrugBank-S1 dataset.
Experimental Results
Experiments conducted on benchmark datasets DrugBank and TWOSIDES demonstrate the efficacy of CBR-DDI. The framework achieves a significant improvement over existing methods, with a 28.7% accuracy increase over baseline models and robust interpretability due to the integration of historical case data.
The ablation studies confirm the importance of both factual drug associations and historical interaction mechanisms. Removing either component results in decreased performance, highlighting the necessity of both factual evidence and deeper mechanistic understanding. Additionally, the hybrid retrieval approach is validated, with balanced semantic and structural reliance yielding optimal results.
Implications and Future Work
The success of CBR-DDI implies broader applications for CBR in augmenting LLM capabilities, especially in domains where historical data play a critical role in decision-making. The interpretability offered by this framework aligns well with clinical practice, where understanding the rationale behind predictions is crucial for adoption in healthcare settings.
However, CBR-DDI's reliance on textual data limits its applicability to molecular-level analysis. Future work could explore integrating molecular structure data to provide richer case representations and deeper mechanistic insights. Such advancements could further refine predictive reliability and extend the framework's utility across more diverse pharmacological challenges.
Conclusion
CBR-DDI represents a significant advancement in DDI prediction, offering a novel integration of LLMs and CBR to produce state-of-the-art results. By leveraging a structured knowledge repository and a dual-layer reasoning approach, the method succeeds in enhancing both the accuracy and interpretability of predictions. This work lays the groundwork for future explorations in combining historical data-driven insights with LLM technologies in the biomedical domain.

