Learning Biomolecular Motion: The Physics-Informed Machine Learning Paradigm (2511.06585v1)
Abstract: The convergence of statistical learning and molecular physics is transforming our approach to modeling biomolecular systems. Physics-informed machine learning (PIML) offers a systematic framework that integrates data-driven inference with physical constraints, resulting in models that are accurate, mechanistic, generalizable, and able to extrapolate beyond observed domains. This review surveys recent advances in physics-informed neural networks and operator learning, differentiable molecular simulation, and hybrid physics-ML potentials, with emphasis on long-timescale kinetics, rare events, and free-energy estimation. We frame these approaches as solutions to the "biomolecular closure problem", recovering unresolved interactions beyond classical force fields while preserving thermodynamic consistency and mechanistic interpretability. We examine theoretical foundations, tools and frameworks, computational trade-offs, and unresolved issues, including model expressiveness and stability. We outline prospective research avenues at the intersection of machine learning, statistical physics, and computational chemistry, contending that future advancements will depend on mechanistic inductive biases, and integrated differentiable physical learning frameworks for biomolecular simulation and discovery.
Sponsored by Paperpile, the PDF & BibTeX manager trusted by top AI labs.
Get 30 days freePaper Prompts
Sign up for free to create and run prompts on this paper using GPT-5.
Top Community Prompts
Explain it Like I'm 14
Overview
This paper is about a new way to teach computers how molecules move and change shape over time. It combines two things:
- Machine learning (ML), which helps computers learn patterns from data.
- Physics, which provides rules that molecules must follow (like conserving energy and obeying thermodynamics).
By mixing both, the goal is to build models that are accurate, trustworthy, and can make good predictions even when we don’t have a lot of data. The paper is a review, meaning it summarizes many recent advances rather than reporting a single experiment.
What questions does the paper try to answer?
The paper focuses on simple but big questions:
- How can we make ML models follow the laws of physics, not just data patterns?
- How can we model molecule movements that happen over very long times (like protein folding), and rare but important events (like a drug binding to a protein)?
- How can we estimate “free energy,” which tells us how likely different molecular shapes are?
- How can we fix the parts that traditional physics models miss (like subtle electronic effects or solvent interactions)?
- What tools and techniques work best, and what are the current challenges?
How do the methods work? (Explained with everyday ideas)
The paper explains several approaches. Think of molecules moving on a huge landscape with hills and valleys:
- Valleys = stable shapes (like a folded protein)
- Hills = barriers that are hard to cross
- Temperature = how much jiggling they do
Here’s how different methods help us learn this landscape and the motion:
Physics-Informed Neural Networks (PINNs)
- Idea: Don’t just fit data—also teach the network the physics rules.
- Analogy: It’s like doing homework where the teacher not only checks your answers but also checks that you used the right formulas.
- Practically: The model gets penalized when it breaks a law (like energy conservation or a known equation).
Operator Learning (DeepONet, Fourier Neural Operator)
- Idea: Learn a “super-calculator” that takes inputs (like boundary conditions or temperatures) and directly outputs solutions to physics problems.
- Analogy: Instead of solving one math problem at a time, learn how to solve whole families of similar problems quickly.
Differentiable Simulation (JAX-MD, TorchMD-Net, TorchSim)
- Idea: Make the simulation itself “differentiable,” so you can adjust model parameters based on how the simulation matches reality, and the computer tells you how to change them.
- Analogy: Like tuning a video game’s physics engine to make it feel realistic, and getting instant feedback on how to fix it.
Hybrid Physics–ML Models
- Idea: Start with a trusted physics model, then add a small ML “correction” to fix missing pieces (like many-body effects or solvent influences).
- Analogy: A chef follows a classic recipe but tweaks it with experience to make it taste better.
- Result: You keep the structure and guarantees of physics but gain flexibility from learning.
Variational Principles and Symmetry
- Variational principles: Nature tends to pick paths or shapes that minimize something (like action or free energy).
- Symmetry: Physics shouldn’t change if you rotate or move a molecule. Models that respect this (called “equivariant”) are more reliable.
- Analogy: A fair game looks the same no matter which direction you look at it.
Stochastic Thermodynamics (Jiggling and Noise)
- Molecules jiggle because of temperature (random noise).
- Models need to handle this noise correctly to stay physically consistent.
- Analogy: Like watching dust specks in the air—they move randomly, but overall their behavior follows rules.
What did the review find, and why does it matter?
This review pulls together many advances and offers a “big picture”:
- Combining physics with ML makes models:
- More accurate (they don’t cheat the rules),
- More stable (they don’t blow up over long simulations),
- More generalizable (they work in new situations).
- These methods help with tough problems:
- Long timescales: Moving from tiny vibrations (trillionths of a second) to slow shape changes (milliseconds or more).
- Rare events: Important but infrequent transitions like protein folding or drug binding.
- Free energy: Estimating which shapes are most likely.
- The “closure problem”:
- Traditional models miss some effects (like subtle electronic changes).
- Hybrid physics–ML models learn corrections that “close the gap” while staying consistent with thermodynamics.
- Tool ecosystem:
- PINNs and operator learning help solve or speed up physics equations.
- Differentiable simulation tools (like JAX-MD, TorchMD-Net 2.0, TorchSim) let you learn directly from simulated trajectories.
- Equivariant graph neural networks respect geometry and symmetry, improving energy and force predictions.
- Trade-offs and challenges:
- Training can be hard and memory-intensive.
- Models must stay stable and physically meaningful.
- Good “inductive biases” (built-in physics-friendly structure) are crucial.
Why should you care?
This approach can speed up and improve:
- Drug discovery: Better models of how drugs bind to proteins.
- Protein engineering: Understanding and controlling how proteins fold.
- Materials science: Predicting how atoms interact in new materials.
By teaching ML to respect physics, we get models that scientists can trust, not just curve fits. That means faster, more reliable discoveries.
What’s the future impact?
The paper argues that progress will come from:
- Building models with strong physics “in their bones” (mechanistic inductive biases).
- Using end-to-end differentiable frameworks where simulations and learning happen together.
- Bridging experiments and simulations, so models can learn directly from lab data while staying physically consistent.
In short, physics-informed machine learning is turning molecular modeling from slow and limited into fast, accurate, and explainable—opening doors for breakthroughs in biology, chemistry, and medicine.
Knowledge Gaps
Knowledge gaps, limitations, and open questions
Below is a consolidated list of concrete gaps and open problems the paper highlights or implies but does not resolve, intended to guide actionable future research:
- Lack of standardized benchmarks for PIML in biomolecular dynamics (datasets, tasks, and metrics) that jointly assess energy conservation, detailed balance, free-energy accuracy, and long-time kinetics.
- Identifiability of drift b(x) and diffusion D(x) from sparse or partial trajectory data: when are these uniquely recoverable, and what minimal data or experimental interventions are needed?
- Hard guarantees vs soft penalties: methods to enforce detailed balance, fluctuation–dissipation (FDT), positivity of entropy production (NESS), and conservative forces by construction rather than via loss terms.
- Training stability and expressiveness of PINNs for stiff, metastable dynamics (Fokker–Planck, committor equations): mitigation of spectral bias, residual weighting pathologies, and poor convergence in barrier-dominated landscapes.
- Systematic handling of periodic boundary conditions and effectively unbounded configuration spaces in PINN/operator-learning setups for molecular systems.
- Generalization of neural operators to new geometries, boundary conditions, and thermodynamic parameters: which inductive biases (E(3) symmetry, conservation, spectral penalties) are necessary and sufficient?
- Quantitative impact of discretization and integrator choice (Ito vs Stratonovich, BAOAB vs VVVR, etc.) on learned SDE dynamics and stationary measures in differentiable simulations.
- Gradient estimation for stochastic dynamics: low-variance, unbiased reparameterizations or likelihood-ratio estimators for Langevin processes at scale; analysis of bias–variance trade-offs.
- Long-horizon learning in differentiable MD: principled algorithms (reversible integrators, multiple shooting, adjoint/implicit gradients) with convergence guarantees and memory/computation bounds.
- Learning position-dependent diffusion tensors D(x): ensuring symmetry, positive-definiteness, FDT consistency with friction, and interpretability; data requirements to avoid overfitting.
- Rare-event kinetics: integrating committor PINNs, transition-path theory, and differentiable MD to estimate MFPTs and rate constants with controlled bias and uncertainty.
- Free-energy estimation losses (Jarzynski/Crooks): variance reduction, bias correction for finite-time nonequilibrium work, and robust training objectives that remain stable under discretization and noise.
- Hybrid closures: certifying that learned corrective forces remain strictly conservative, extensive, and thermodynamically consistent; detecting and preventing hidden nonconservative artifacts in GNN architectures.
- Transferability across thermodynamic state points (T, ionic strength, solvent models): training protocols and constraints that guarantee consistent ensembles and kinetics under state changes.
- Mechanistic interpretability of learned corrections: attributing Eθ to physical effects (polarization, many-body dispersion, solvent-mediated forces), and developing diagnostics/probes to validate these attributions.
- Uncertainty quantification: Bayesian/ensemble PIML and differentiable MD with calibrated uncertainties on energies, free energies, and kinetic observables; propagation to experimental predictions.
- Data efficiency and active learning: minimal trajectory lengths/coverage needed to recover slow modes; acquisition strategies that adaptively sample states to reduce uncertainty in dynamics and FES.
- Multi-fidelity learning: principled weighting and debiasing across QM, MM, CG, and experimental constraints to avoid double counting while preserving thermodynamic consistency.
- CV discovery benchmarks: standardized suites comparing VAMP/TICA/VAMPnets and equivariant encoders vs hand-crafted CVs on complex systems with out-of-sample tests for kinetics and FES.
- Scalability to large biomolecular assemblies: sparse/equivariant architectures and neighbor-list AD that preserve differentiability; multi-GPU/TPU strategies without sacrificing stability or correctness.
- Differentiable experimental forward models: accurate, noise-aware, and differentiable likelihoods for SAXS, RDC/NOE, cryo-EM, and scattering observables; identifiability under partial observability.
- Robust bridges to established MD engines: low-overhead, differentiable interfaces (LAMMPS/OpenMM) with consistent integrator semantics and gradients; validation against reference solvers.
- Managing discretization artifacts in end-to-end learning: correction schemes or shadow-work accounting to maintain correct stationary distributions and kinetics under finite time steps.
- Validation in nonequilibrium regimes: standardized NESS testbeds with known entropy production to evaluate Crooks/Jarzynski-constrained learning and path-measure consistency.
- Operator-learning surrogates for electrostatics/diffusion in heterogeneous media: handling charge singularities, dielectric discontinuities, and enforcing integral constraints/boundary flux conservation.
- Holonomic constraints in differentiable MD: stable, differentiable SHAKE/RATTLE variants with accurate gradients and minimal drift over long trajectories.
- Safety and extrapolation control: certifiable bounds (e.g., Lipschitz constraints, energy floors/upper bounds) to prevent catastrophic extrapolation in unseen chemistries or configurations.
- Automated and principled loss balancing: adaptive schemes with theoretical backing to set λ-weights between data, residual, boundary, and multi-scale objectives and avoid domination by any single term.
- Reweightability and kinetics preservation under learned biasing/control: conditions under which enhanced-sampling biases remain exactly reweightable and do not corrupt dynamical observables.
- Standard datasets and protocols for reproducibility: open trajectories, PDE problem sets, evaluation scripts, and seeds for fair comparison across PINNs, operator learning, and differentiable MD frameworks.
- Comprehensive error auditing: separating model, discretization, and sampling errors; tools to attribute discrepancies in observables to specific sources in hybrid physics–ML pipelines.
Practical Applications
Immediate Applications
Below are actionable use cases that can be deployed now using the frameworks, tools, and methods discussed in the paper. Each item lists sector(s), a concrete workflow or product concept, and key assumptions or dependencies that influence feasibility.
- Physics-informed free-energy estimation and binding affinity ranking for drug discovery
- Sectors: healthcare/biopharma, software
- Workflow/product: A SaaS tool that runs hybrid physics–ML simulations to compute ΔG and rank ligands, combining baseline force fields (e.g., AMBER/CHARMM) with learnable closures Eθ trained via differentiable MD (TorchMD-Net 2.0, JAX-MD, TorchSim). Integrate ensemble calibration against experimental observables (SAXS, NMR) and reweight results using Jarzynski/Crooks identities to ensure thermodynamic consistency.
- Assumptions/dependencies: Reliable baseline force field; GPU capacity for batched trajectories; validated mapping from experimental data to ensemble constraints; correct enforcement of fluctuation–dissipation theorem (FDT) and detailed balance in equilibrium runs.
- Conformational landscape mapping and rare-event analysis for proteins and complexes
- Sectors: healthcare/biopharma, academia
- Workflow/product: Use VAMP/TICA objectives and VAMPnets to learn slow collective variables; drive enhanced sampling (e.g., metadynamics, adaptive sampling) to characterize folding pathways, allosteric transitions, and on/off-target binding. Equip with equivariant GNN encoders to preserve E(3) symmetries.
- Assumptions/dependencies: Sufficient trajectory coverage; careful handling of boundary/initial conditions; validation of metastable state decomposition; appropriate lag times in operator learning.
- Operator-learning surrogates for biomolecular PDEs (electrostatics, diffusion, committor)
- Sectors: biopharma/materials, software, academia
- Workflow/product: Build resolution-invariant, fast surrogates for Poisson–Boltzmann or Fokker–Planck solves using DeepONet/FNO and PINN residual losses (DeepXDE, PhysicsNeMo). Deploy as APIs in docking, pKa prediction, and transport simulations to accelerate parameter sweeps and screening.
- Assumptions/dependencies: Robust generalization across geometries and boundary conditions; representative training distributions; spectral/physics regularization during training to prevent drift off the conservation manifold.
- Experiment-in-the-loop refinement of potentials and ensembles
- Sectors: biopharma, academia
- Workflow/product: End-to-end differentiable pipelines that backpropagate from SAXS/NMR/cryo-EM objectives to hybrid potentials Ephys + Eθ using TorchMD-Net 2.0 or TorchSim. Multi-objective losses couple microscopic force matching to mesoscopic ensemble fidelity and kinetic targets.
- Assumptions/dependencies: Accurate forward models from structure/trajectory to experimental observables; robust treatment of measurement noise and uncertainty; memory-managed training (checkpointing, multiple shooting) for long rollouts.
- Coarse-grained (CG) closures that retain detailed balance and transferability
- Sectors: biopharma/materials, academia
- Workflow/product: Learn CG force corrections and spatially varying diffusion Dθ via differentiable MD in the overdamped regime, enforcing FDT and equilibrium consistency. Use to simulate large macromolecular assemblies, polymers, and soft matter at long timescales.
- Assumptions/dependencies: High-quality AA reference data; friction/diffusion fields calibrated to environment; careful thermodynamic validation across state points.
- Differentiable enhanced sampling and control protocol optimization
- Sectors: biopharma, academia
- Workflow/product: Optimize biasing schedules (e.g., time-dependent metadynamics or umbrella windows) by differentiating through the integrator to minimize mean first passage times (MFPT) while preserving reweightability. Use TorchMD-Net 2.0 or TorchSim with stochastic integrators and reparameterized noise.
- Assumptions/dependencies: Stable gradients for stochastic dynamics; accurate estimators for path-measure ratios; safeguards against training instabilities in stiff systems.
- PIML MLOps and GPU throughput tooling
- Sectors: software/HPC
- Workflow/product: Adopt TorchSim’s AutoBatching to pack heterogeneous systems for concurrent MD integration; standardize experiment tracking and checkpointing for PINN/neural-operator training at scale. Provide a “physics-consistency CI” that checks energy conservation, detailed balance, and entropy production constraints on every model release.
- Assumptions/dependencies: Modern GPU hardware; robust data loaders and neighbor-list generation; organization-wide adoption of physics-aware model validation.
- Educational modules and reproducible labs in physics-informed ML
- Sectors: education/academia
- Workflow/product: Course kits with JAX-MD/TorchMD-Net notebooks that teach energy landscapes, Langevin dynamics, PINNs, and neural operators via hands-on exercises. Include benchmarking datasets and reference solutions for committor fields and diffusion.
- Assumptions/dependencies: Access to GPUs or CPU fallback; curated examples that fit classroom constraints; instructor familiarity with autodiff and PDE residuals.
- Mechanistic documentation for regulatory submissions (in silico support)
- Sectors: policy/regulation, biopharma
- Workflow/product: Provide physics-informed audit trails—model cards documenting conservation checks, equilibrium validation, and experimental reweighting—in support of computational claims (e.g., binding predictions) in preclinical packages.
- Assumptions/dependencies: Clear guidance from regulators on acceptable in silico evidence; internal SOPs for physics-consistency reporting; cross-validation with experimental data.
Long-Term Applications
These use cases require further method development, scaling, or integration across disciplines, but are directly motivated by the paper’s methods and vision.
- Autonomous, closed-loop molecular design with physics-informed generative models
- Sectors: biopharma/materials, software, robotics
- Workflow/product: Couple Boltzmann generators or score-based flows regularized by PINN/PDE constraints with differentiable MD for design–evaluate–learn cycles. Robotic platforms synthesize and test candidates; models update via experiment-in-the-loop training to enforce thermodynamic and kinetic plausibility.
- Assumptions/dependencies: Robust generative priors that maintain detailed balance under constraints; scalable differentiable simulation; lab automation and data integration; strong uncertainty quantification.
- Personalized molecular simulations for precision medicine
- Sectors: healthcare
- Workflow/product: Patient-specific ensembles and kinetics for mutant proteins or variant receptors, predicting stability changes, binding shifts, and allosteric effects. Use hybrid closures to adapt baseline physics to individual molecular contexts.
- Assumptions/dependencies: High-quality patient-specific structural models; clinically actionable turnaround times; validated pipelines linking simulation outputs to treatment decisions.
- Multiscale “digital twins” of cellular environments
- Sectors: healthcare/biotech, academia, policy
- Workflow/product: Integrate operator-learning surrogates (transport/reaction-diffusion) with CG dynamics and stochastic thermodynamics to model intracellular kinetics and transport under physiological perturbations. Apply to mechanism-of-action studies and safety assessments.
- Assumptions/dependencies: Reliable cross-scale coupling (AA→CG→continuum); data assimilation from imaging and omics; governance frameworks for model validation in decision-making.
- Real-time screening platforms powered by neural operator surrogates
- Sectors: biopharma/materials, cloud software
- Workflow/product: Resolution-invariant FNO/DeepONet services for families of biomolecular PDEs (electrostatics, solvent response, Fokker–Planck kinetics), enabling massive parameter sweeps (temperature, boundary charges, ligand poses) with physics-regularized inference.
- Assumptions/dependencies: Training across diverse geometries and state points; drift detection and correction; standardized benchmarks for out-of-distribution generalization.
- Physics-informed closures for complex soft-matter and energy systems
- Sectors: energy, advanced materials, consumer products
- Workflow/product: Hybrid models that recover polarization, many-body effects, and solvent coupling for electrolytes, membranes, and polymers (e.g., battery electrolytes, filtration materials). Use differentiable simulation to fit to transport and rheology observables.
- Assumptions/dependencies: Domain-specific datasets (transport coefficients, conductivity, scattering); scalable training for large heterogeneous systems; robust treatment of anisotropic diffusion Dθ.
- Standardized regulatory frameworks for physics-informed ML in drug development
- Sectors: policy/regulation
- Workflow/product: Guidance documents and validation protocols that recognize physics-consistency metrics (energy conservation, detailed balance, Jarzynski/Crooks compliance) as part of model qualification for in silico evidence.
- Assumptions/dependencies: Consensus on validation criteria; cross-industry/agency collaboration; case studies demonstrating predictive value and reproducibility.
- Curriculum and workforce development for integrated ML–physics
- Sectors: education/academia/industry
- Workflow/product: Degree tracks and micro-credentials focused on PIML, differentiable simulation, and operator learning; capstones that deploy PINNs/DP in real biomolecular problems; open benchmarks and community datasets.
- Assumptions/dependencies: Institutional buy-in; sustained funding; accessible tooling and documentation.
- Generalization of stochastic thermodynamics constraints to AI systems beyond molecules
- Sectors: software/AI research (potentially finance/robotics)
- Workflow/product: Apply pathwise consistency (entropy production bounds, reversibility) to stabilize long-horizon learned dynamics (e.g., robot control under uncertainty, certain financial microstructure simulators). Use SDE-PINNs for drift/diffusion identification with physical-style constraints.
- Assumptions/dependencies: Careful mapping of physical constraints to domain-specific analogs; validation that thermodynamic-style priors improve performance; ethical considerations in sensitive domains.
Cross-cutting assumptions and dependencies
- Data quality and coverage: High-fidelity QM/AA data and experimental observables are needed to train closures and validate ensembles/kinetics.
- Computational resources: Differentiable simulation is GPU-intensive; memory and stability require checkpointing, multiple shooting, or adjoint/implicit gradients.
- Physical correctness: Maintaining FDT, energy conservation, detailed balance, and thermodynamic consistency is critical; architecture-level inductive biases (equivariance, conservative heads) often outperform soft penalties alone.
- Stability and expressiveness: PINN/operator solutions can be sensitive to residual weighting; differentiable MD can be unstable for stiff systems; careful hyperparameter tuning and integrator selection are required.
- Generalization: Operator-learning surrogates must handle geometry/parameter shifts; physics-aware regularization and spectral penalties help, but standardized benchmarks are needed.
- Validation and governance: For policy/clinical uses, rigorous model documentation, uncertainty quantification, and reproducibility standards are essential.
Glossary
- ab initio: From first-principles quantum mechanics (no empirical parameters), often used as a gold standard for accuracy. "achieving ab initio accuracy at classical MD cost."
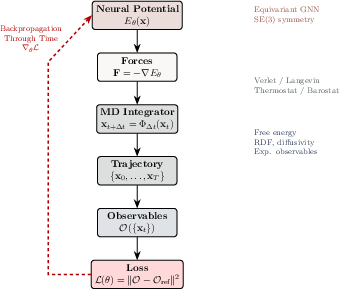
- Backpropagation through time (BPTT): Gradient-based training through unrolled time steps of a dynamical system. "Gradients ... are obtained by backpropagation through time (BPTT), with memory requirements managed via checkpointing or implicit differentiation."
- Behler-Parrinello neural network potential: A symmetry-aware neural potential that decomposes molecular energy into atomic contributions derived from local environments. "The Behler-Parrinello neural network potential"
- Boltzmann distribution: The equilibrium probability distribution over states weighted by energy at temperature T. "equilibrium probabilities follow the Boltzmann distribution."
- Boltzmann Generators: Generative models trained to sample configurations according to Boltzmann weights for unbiased equilibrium sampling. "Generative formulations such as Boltzmann Generators try to achieve this by embedding directly within the training objective"
- Boltzmann machine: A stochastic energy-based neural network inspired by statistical physics used for learning probability distributions. "the Boltzmann machine"
- Canonical ensembles: Statistical ensembles at fixed particle number, volume, and temperature (NVT), governed by Boltzmann weights. "canonical ensembles in thermodynamics"
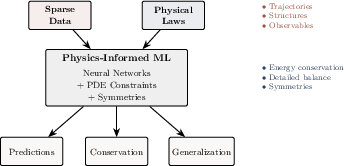
- Closure problem (biomolecular closure problem): The challenge of modeling unresolved interactions omitted by coarse or empirical models while preserving physical consistency. "the biomolecular analogue of the closure problem lies in representing unresolved interactions that classical potentials neglect"
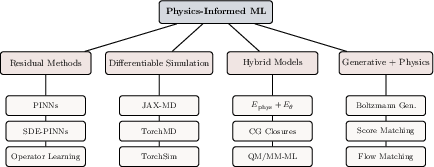
- Coarse-grained (CG): A reduced-resolution modeling approach that groups atoms into larger beads to accelerate simulation. "Coarse-grained (CG) closures:"
- Committor function: The probability that a trajectory starting at a configuration reaches one state before another; solves a backward Kolmogorov equation. "or committor function"
- Crooks’ theorem: A fluctuation relation connecting probabilities of forward and reverse nonequilibrium trajectories. "Crooksâ theorem relates the probabilities of forward and reverse paths, "
- Deep Potential Molecular Dynamics: A framework that learns many-body potentials from quantum data to run accurate, scalable MD simulations. "Deep Potential Molecular Dynamics \cite{Zhang2018-zv} extended this principle to scalable many-body systems"
- DeepONet: A neural operator architecture that learns mappings between functions using branch and trunk networks. "DeepONet, which composes branch and trunk nets to achieve universal approximation of nonlinear operators"
- Detailed balance: A microscopic reversibility condition ensuring equilibrium dynamics have no net probability flux between states. "including energy conservation, detailed balance, and thermodynamic consistency."
- Differentiable physics: Simulation frameworks embedded in computation graphs so that gradients can flow through numerical solvers. "In differentiable physics (DP) frameworks"
- E(3)-equivariant graph neural networks: Models that preserve 3D Euclidean symmetries (rotations, translations, reflections) for physically consistent predictions. "E(3)-equivariant graph neural networks"
- Fluctuation theorems: Exact results relating nonequilibrium fluctuations to equilibrium thermodynamics and free-energy differences. "Fluctuation theorems provide exact constraints"
- Fluctuation-dissipation theorem (FDT): The principle linking thermal noise strength to dissipative friction to maintain equilibrium. "satisfying the fluctuation-dissipation theorem (FDT)."
- Fokker-Planck equation: A PDE describing the time evolution of probability densities under stochastic dynamics. "The corresponding Fokker-Planck equation (utilized to represent the temporal evolution of the probability density function for systems influenced by stochastic variations)"
- Fourier Neural Operator (FNO): A neural operator that learns resolution-invariant solutions to PDE families via spectral transforms. "the Fourier Neural Operator (FNO) learns resolution-invariant mappings via spectral transforms"
- Free-energy surface (FES): The potential of mean force over collective variables that encodes thermodynamics and kinetics. "The free-energy surface (FES) "
- Hamiltonian and Lagrangian neural networks: Architectures that learn energy or action functionals whose derivatives yield physics-consistent equations of motion. "Hamiltonian and Lagrangian neural networks learn energy or action functionals"
- Hopfield network: An energy-based recurrent neural network with attractor dynamics inspired by spin-glass models. "the Hopfield network"
- Jarzynski equality: An identity relating the exponential average of nonequilibrium work to equilibrium free-energy differences. "the Jarzynski equality "
- Langevin dynamics: Stochastic equations of motion modeling thermalized systems with friction and noise. "For molecular systems evolving under Langevin dynamics"
- Many-body effects: Interactions that involve three or more particles simultaneously, beyond pairwise forces. "many-body effects"
- Mean first-passage time (MFPT): The average time for a stochastic process to reach a target state for the first time. "$\big[\mathrm{MSE}(\mathrm{VACF}),\,|\!D_\theta-D_{\mathrm{ref}|,\,|\mathrm{MFPT}_\theta-\mathrm{ref}|\Big]$"
- Nonequilibrium steady state (NESS): A stationary state with sustained fluxes and nonzero entropy production under driving. "driven steady states (NESS)"
- Normalizing-flow models: Invertible neural generative models with tractable densities used for sampling and free-energy estimation. "normalizing-flow models"
- Onsager-Machlup action: A functional whose minimizers characterize the most probable paths of stochastic processes. "Onsager-Machlup action (a function that encapsulates the dynamics of a continuous stochastic process)"
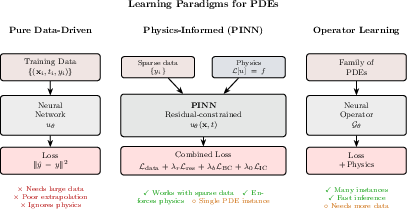
- Operator learning: Learning mappings from input functions/conditions to solution fields, amortizing PDE solves. "Operator-learning architectures approximate directly"
- Overdamped regime: The high-friction limit where inertia is negligible and dynamics reduce to positional diffusion. "In the high-friction (overdamped) regime"
- Partition function: The normalization integral over all configurations that determines equilibrium thermodynamics. "forming an equilibrium ensemble defined by the partition function"
- Physics-Informed Machine Learning (PIML): ML approaches that integrate physical laws and constraints into training for accuracy and consistency. "Physics-informed machine learning (PIML) offers a systematic framework that integrates data-driven inference with physical constraints"
- Physics-Informed Neural Networks (PINNs): Neural networks trained by penalizing PDE residuals, boundary, and initial condition violations. "Physics-Informed Neural Networks (PINNs), introduced by Raissi, Perdikaris, and Karniadakis"
- Poisson-Boltzmann: A continuum electrostatics PDE used to model ionic screening around biomolecules. "Poisson-Boltzmann"
- Reaction-diffusion kinetics: PDE-based models capturing species diffusion coupled with chemical reactions over space and time. "reaction-diffusion kinetics"
- Smoluchowski: The Smoluchowski equation/model for diffusive (overdamped) dynamics in configuration space. "Smoluchowski"
- Transfer operator: The linear operator that propagates probability densities or observables in time, central to kinetics. "the transfer operator"
- Underdamped regime: Dynamics where inertia matters and both positions and velocities evolve with friction and noise. "the underdamped hybrid Langevin equation"
- VAMP (Variational Approach for Markov Processes): A variational framework to learn slow collective variables by approximating leading transfer-operator eigenfunctions. "The variational approach for Markov processes (VAMP)"
- VAMPnets: Neural architectures implementing VAMP to learn slow modes and metastable states from trajectories. "VAMPnets \cite{Mardt2018-th}"
- Velocity autocorrelation function (VACF): A time correlation function used to characterize kinetic properties like diffusion. "$\big[\mathrm{MSE}(\mathrm{VACF}),\,|\!D_\theta-D_{\mathrm{ref}|,\,|\mathrm{MFPT}_\theta-\mathrm{ref}|\Big]$"
Collections
Sign up for free to add this paper to one or more collections.