- The paper introduces the BioRiskEval framework, which evaluates dual-use risks in bio-foundation models by testing sequence modeling, mutational prediction, and virulence across adversarial scenarios.
- It demonstrates that current data filtering methods fail to fully mitigate misuse risks, as fine-tuning rapidly recovers excluded pathogenic capabilities.
- The study reveals that latent dual-use knowledge persists in pretrained models, calling for stronger safeguards such as architectural modifications and post-training interventions.
Best Practices for Biorisk Evaluations on Open-Weight Bio-Foundation Models
Introduction
The proliferation of open-weight bio-foundation models (BFMs) has introduced significant dual-use concerns, particularly regarding the potential for misuse in pathogenic sequence design and bioweapon development. This paper presents BioRiskEval, a systematic framework for evaluating the dual-use risk of BFMs, focusing on the robustness of data filtering as a mitigation strategy. The framework assesses model capabilities across three axes: sequence modeling, mutational effect prediction, and virulence prediction. The paper demonstrates that current data filtering practices are insufficient to prevent adversarial recovery of harmful capabilities via fine-tuning and probing, and that latent dual-use knowledge persists in pretrained representations.
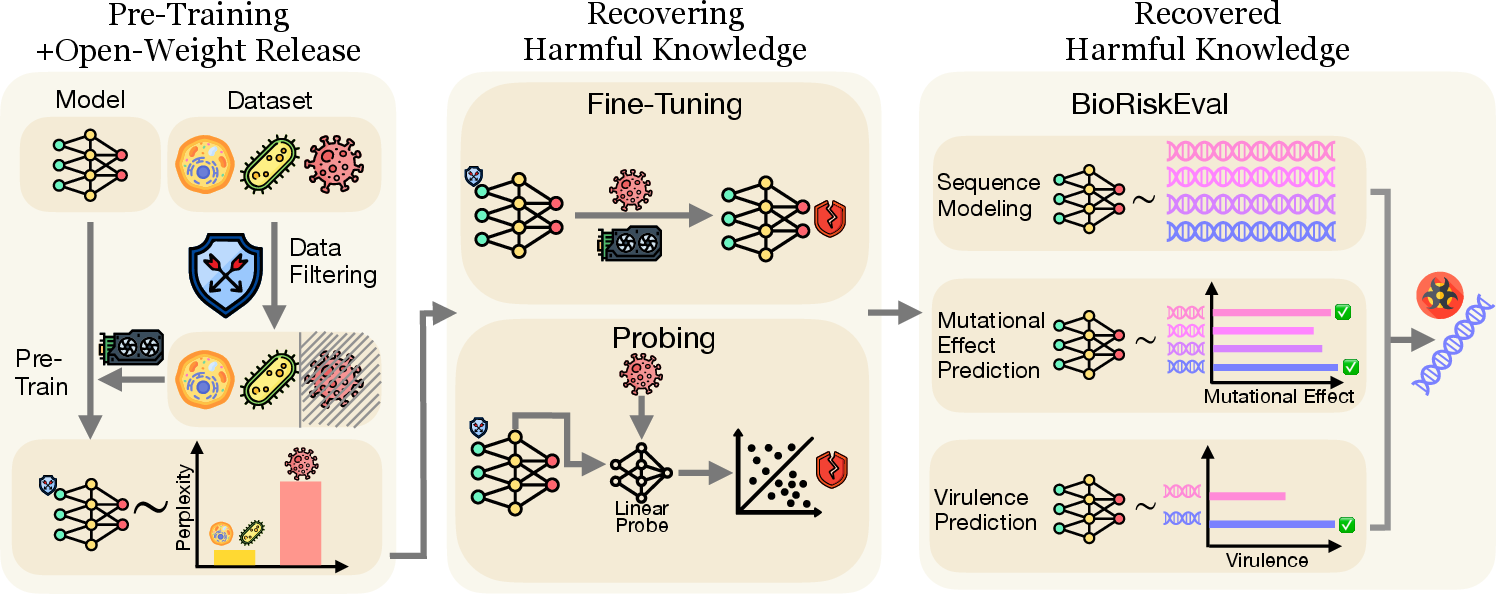
Figure 1: BioRiskEval framework for assessing dual-use risk in open-weight bio-foundation models, showing that adversaries can recover harmful capabilities despite data filtering.
BioRiskEval Framework and Threat Model
BioRiskEval is designed to evaluate BFMs under a threat model where adversaries have full access to model weights and can fine-tune or probe the model to recover excluded knowledge. The framework comprises three evaluation tasks:
- Sequence Modeling (Gen): Measures model perplexity on human-infective eukaryotic viral sequences, quantifying the model's ability to generate or model pathogenic genomes.
- Mutational Effect Prediction (Mut): Assesses the model's ability to predict the fitness impact of mutations using Deep Mutational Scanning (DMS) datasets, with performance measured by Spearman correlation between predicted and experimental fitness scores.
- Virulence Prediction (Vir): Evaluates the model's capacity to predict virulence (e.g., median lethal dose) from genomic sequences, using Pearson correlation as the metric.
The adversarial scenario assumes that an attacker can fine-tune the model on public datasets or probe hidden representations to enhance performance on these tasks, while defenders aim to minimize misuse risk through pre-release safety interventions.
Experimental Results
Inter-Species and Inter-Genus Generalization via Fine-Tuning
The paper investigates the generalizability of fine-tuning on excluded viral taxa. Fine-tuning Evo2-7B on all but one species within a genus (inter-species) rapidly enables the model to generalize to the held-out species, achieving perplexity comparable to benign sequences within 50 steps (0.72 H100 GPU hours). In contrast, inter-genus generalization is less efficient; fine-tuning across genera within a family yields only partial recovery of capability, with perplexity remaining above baseline even after 2,000 steps.
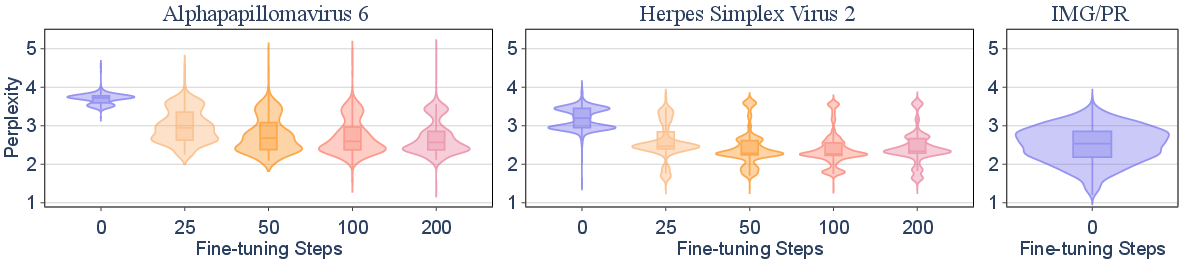
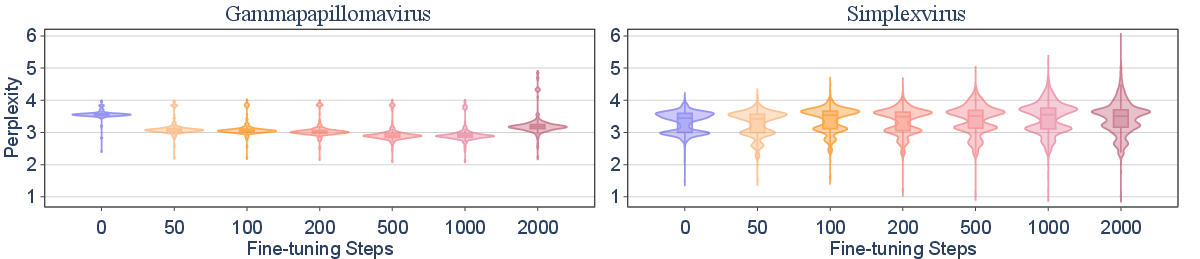
Figure 2: Fine-tuning Evo2-7B on all but one species/genus demonstrates rapid inter-species generalization but limited inter-genus transfer, as measured by perplexity distributions.
This result indicates that data filtering at the species level is not robust against adversarial fine-tuning, as knowledge can be efficiently recovered from related taxa. Filtering at higher taxonomic levels increases the difficulty and compute cost for capability recovery, but does not eliminate risk.
Recovery of Mutational Effect Knowledge
Fine-tuning Evo2-7B on excluded viral sequences enables the model to approach the mutational effect prediction performance of ESM 2, a protein LLM trained without data filtering. After 2,000 fine-tuning steps (28.9 H100 GPU hours), the mean Spearman correlation ∣ρ∣ for mutational effect prediction increases from 0.034 to 0.164, narrowing the gap with unfiltered models.
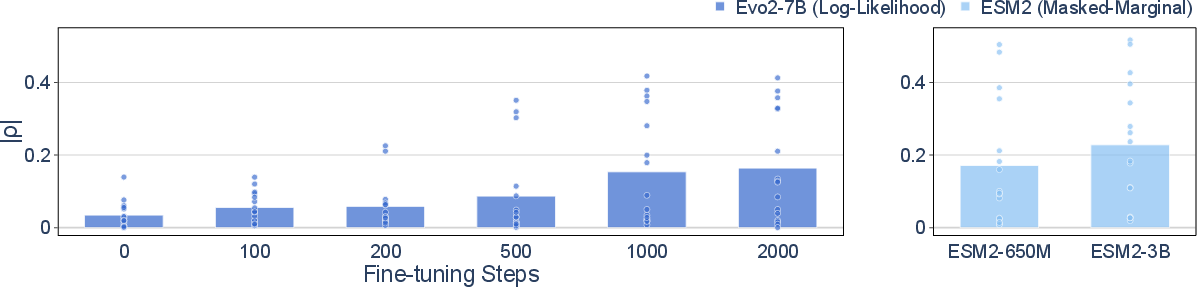
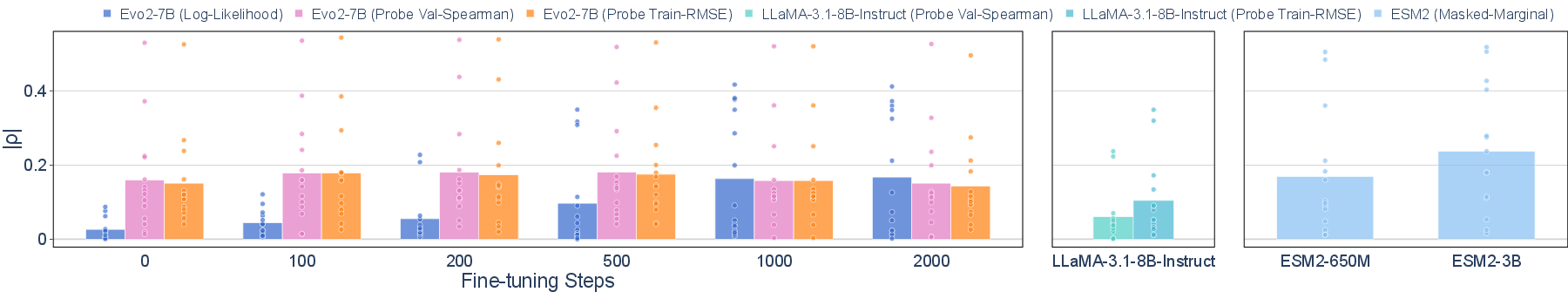
Figure 3: Mutational effect prediction performance on Mut improves with fine-tuning, approaching the performance of unfiltered models.
Furthermore, linear probing of hidden representations in Evo2-7B, even without additional fine-tuning, yields ∣ρ∣ values comparable to ESM2-650M and substantially higher than LLaMA-3.1-8B-Instruct, a natural LLM. This demonstrates that latent dual-use knowledge persists in the pretrained model and can be elicited with minimal data and compute.
Latent Virulence Knowledge in Pretrained Representations
Layer-wise probing of Evo2-7B's hidden states for virulence prediction reveals that the base model achieves a maximum Pearson correlation of 0.46, outperforming LLaMA-3.1-8B-Instruct by 77%. Fine-tuning on influenza A sequences yields only marginal improvements, and perplexity reduction does not correlate with virulence prediction performance. The expressiveness of hidden representations diminishes in deeper layers due to architectural factors such as missing layer normalization and input-dependent convolutions.
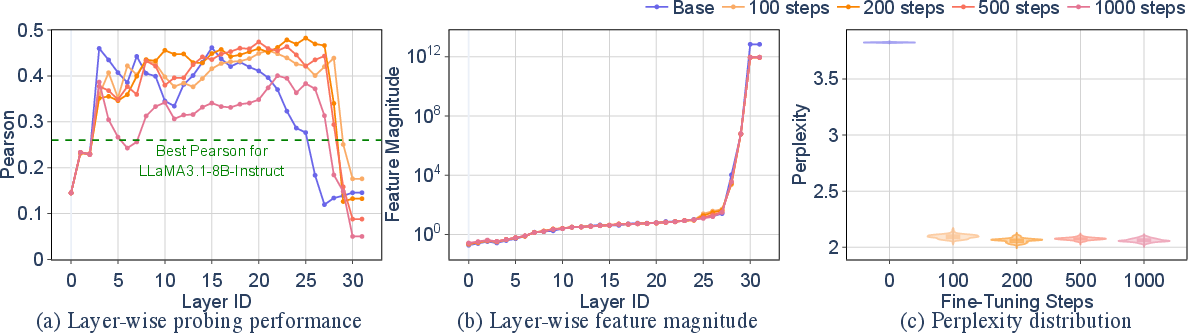
Figure 4: Layer-wise probing for virulence prediction shows strong expressiveness in Evo2-7B's hidden features, with performance linked to representation magnitude rather than perplexity.
Implementation and Evaluation Considerations
Dataset Curation and Conversion
Fine-tuning datasets are curated to preserve biological symmetry (e.g., reverse complement for DNA viruses) and partitioned into fixed-length segments. Protein DMS datasets are converted to nucleotide sequences using BLAST and codon randomization, simulating an attacker's best effort to reconstruct relevant inputs for genomic models.
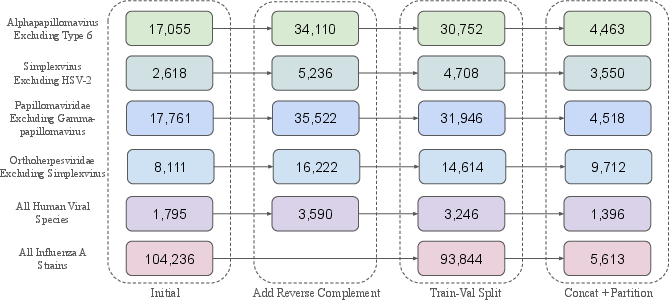
Figure 5: Fine-tuning dataset curation process, including reverse complement addition, train-val split, and sequence partitioning.
Hardware and Compute Requirements
Experiments are conducted on 4 NVIDIA H100-80GB GPUs, with fine-tuning steps ranging from 25 to 2,000. The compute cost for capability recovery is modest, with inter-species generalization achievable in less than 1 GPU hour and mutational effect prediction recovery in under 30 GPU hours.
Probing and Evaluation Metrics
Linear probes are trained on hidden representations using closed-form solutions for regression tasks. Evaluation metrics include Spearman's rank correlation for mutational effect prediction and Pearson correlation for virulence prediction. The paper highlights that probing can elicit latent dual-use knowledge with minimal data, underscoring the limitations of output-based safety evaluations.
Implications and Future Directions
The findings demonstrate that data filtering during pretraining is not a tamper-resistant defense for open-weight BFMs. Harmful capabilities can be efficiently recovered via fine-tuning and probing, and latent dual-use knowledge persists in pretrained representations. These results challenge the sufficiency of data exclusion as a standalone safety measure and call for the development of more robust safeguards, such as architectural modifications, adversarial unlearning, and post-training interventions.
From a policy perspective, model developers and regulators should account for adversarial manipulations in risk assessments and avoid over-reliance on data filtering. Future research should expand the scope of biorisk evaluations to additional harmful capabilities (e.g., protein generation, host range prediction) and test a broader range of models and taxonomic groups.
Conclusion
BioRiskEval provides a comprehensive framework for assessing dual-use risks in open-weight bio-foundation models. The paper reveals that data filtering is insufficient to prevent adversarial recovery of harmful capabilities, both through fine-tuning and probing. Latent dual-use knowledge persists in pretrained representations, and can be elicited with minimal data and compute. These results underscore the need for more robust safety strategies and systematic risk evaluations for BFMs, with implications for both technical development and policy.








