- The paper presents a robust multi-omics aging clock leveraging LightGBM, achieving high predictive accuracy with Pearson’s R up to 0.95.
- It uncovers distinct aging archetypes via unsupervised clustering and identifies key biomarkers from transcriptome, metabolome, lipidome, and microbiome data.
- The study highlights sex-specific molecular waves and establishes strong links between microbiome AgeAccel scores and clinical disease risks for personalized intervention.
Phenome-Wide Multi-Omics Integration Reveals Distinct Archetypes of Human Aging
Introduction
This paper presents a comprehensive phenome-wide analysis of human aging by integrating multi-omics data—transcriptomics, metabolomics, lipidomics, and microbiome profiles—collected from a large, deeply phenotyped cohort (Human Phenotype Project, HPP) of over 10,000 adults aged 30–70 years. The central objective is to develop and validate a multi-omics aging clock capable of robustly predicting biological age, health outcomes, and disease risk, while uncovering distinct molecular subtypes and trajectories of aging. The work leverages advanced nonlinear machine learning frameworks, notably LightGBM, to model the complex, nonlinear dynamics of aging across biological layers.
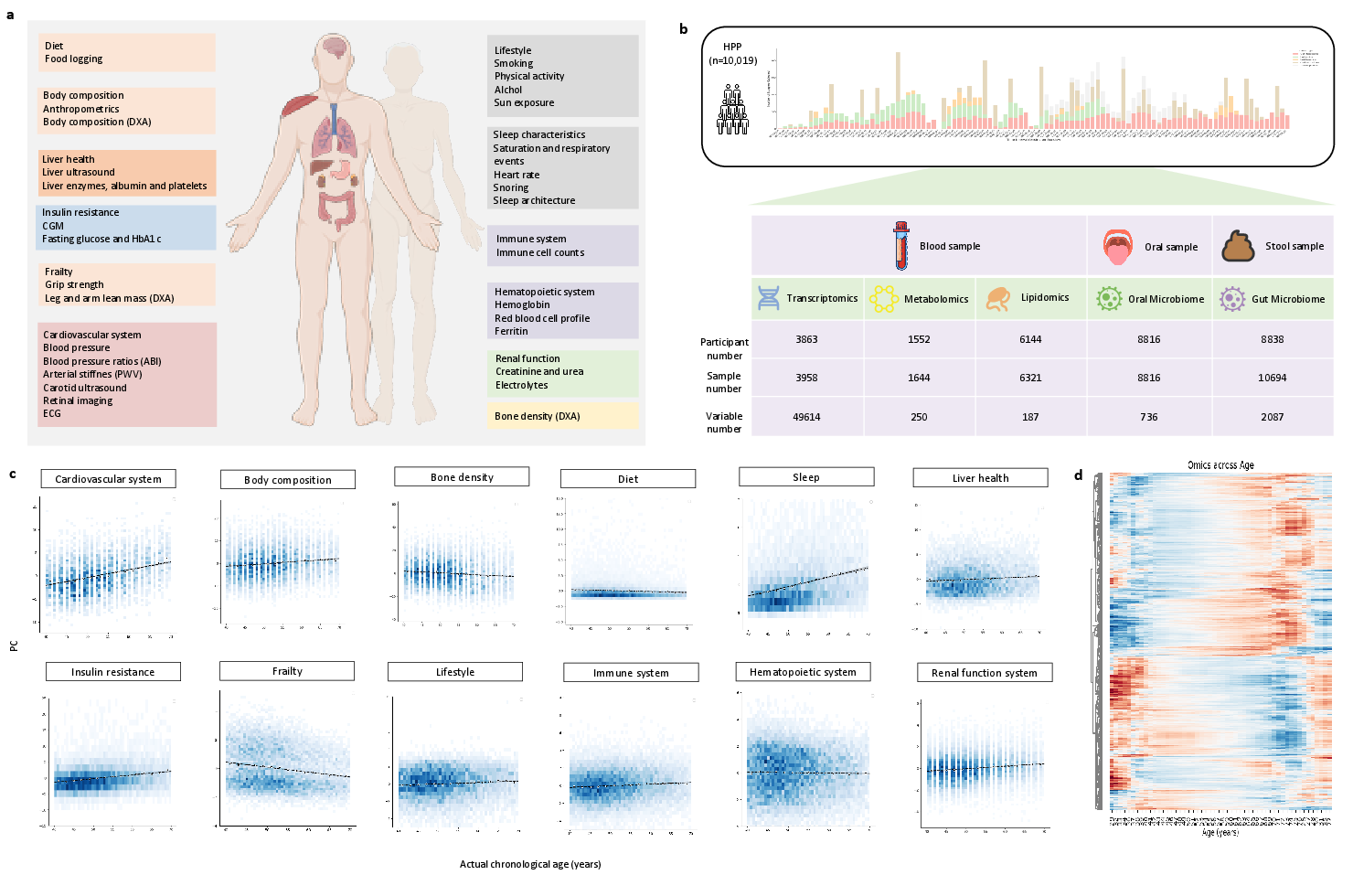
Figure 1: System-wide multi-omics and phenotypic data collection in the HPP cohort, illustrating the breadth of molecular and physiological features profiled across 12 body systems.
Cohort Design and Data Acquisition
The HPP cohort comprises ethnically diverse, predominantly healthy individuals with extensive longitudinal follow-up. Biological samples (blood, stool, oral swabs) were collected at baseline and processed for high-dimensional multi-omics profiling, yielding over 52,000 molecular features and nearly 1013 data points. The dataset encompasses 12 system-level categories, including cardiovascular, metabolic, immune, and lifestyle domains, enabling a holistic assessment of aging phenotypes.
Principal component and correlation analyses demonstrate significant age dependency across most systemic indicators, with cardiovascular and sleep measures showing the strongest associations. Molecular features exhibit pronounced nonlinear changes with age, underscoring the necessity of nonlinear modeling approaches.
Construction and Validation of Multi-Omics Aging Clocks
Aging clocks were constructed for each omics layer using LASSO, Elastic Net, and LightGBM, with LightGBM consistently outperforming linear models in predictive accuracy and robustness. Feature selection employed a two-stage process: Spearman correlation filtering (FDR-adjusted p<0.05) followed by SHAP-based importance ranking.
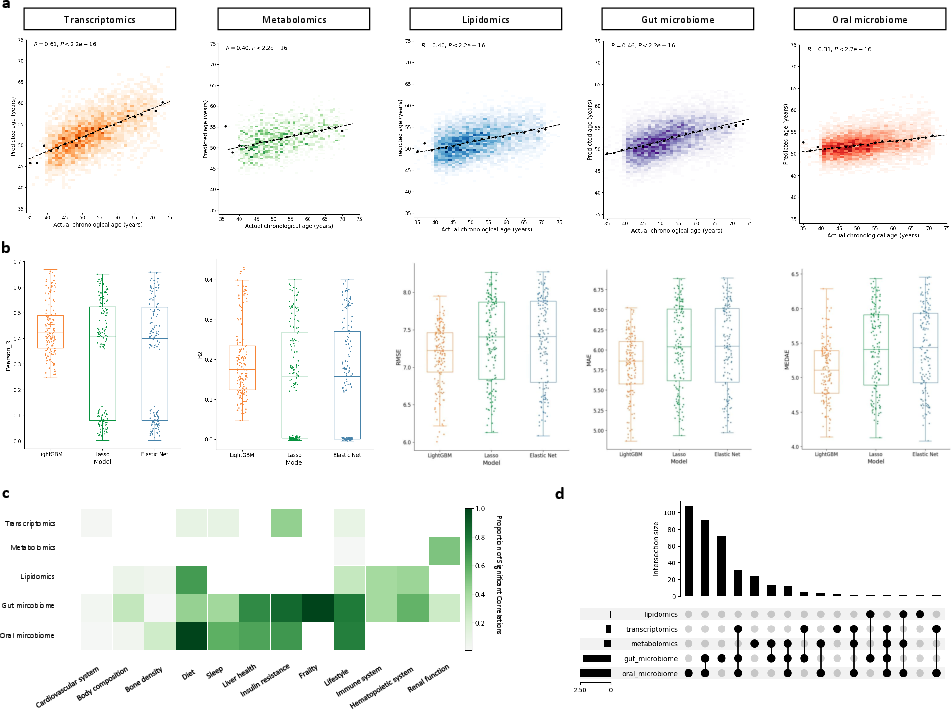
Figure 2: Internal validation of multi-omics aging clocks, showing high Pearson correlation between predicted and actual age, robustness across resampling, and systemic relevance of AgeAccel scores.
The multi-omics clocks exhibit strong correlations with chronological age (Pearson’s R up to 0.95 for transcriptomics), low median absolute errors, and high R2 values. Age acceleration (AgeAccel) scores, defined as residuals from regressing biological age on chronological age, reveal divergent associations across physiological systems. Microbiome-based clocks (gut and oral) display the broadest systemic relevance, with significant links to liver health, frailty, bone density, and lifestyle factors. Molecular clocks (transcriptome, metabolome, lipidome) show more circumscribed associations, e.g., transcriptomic AgeAccel with insulin resistance and metabolomic AgeAccel with renal function.
Analysis of biomarker overlap and specificity demonstrates that each omics clock captures unique and shared biological signals, with oral microbiome AgeAccel retaining the largest unique subset of associated parameters.
Molecular Archetypes and Trajectories of Aging
Unsupervised clustering of integrated omics data identifies distinct aging archetypes: accelerated and decelerated aging. Accelerated aging is characterized by early, large-scale disruption of molecular and microbial homeostasis, with significant changes manifesting as early as age 50. Decelerated aging exhibits delayed and blunted molecular changes, with stability persisting into later life.
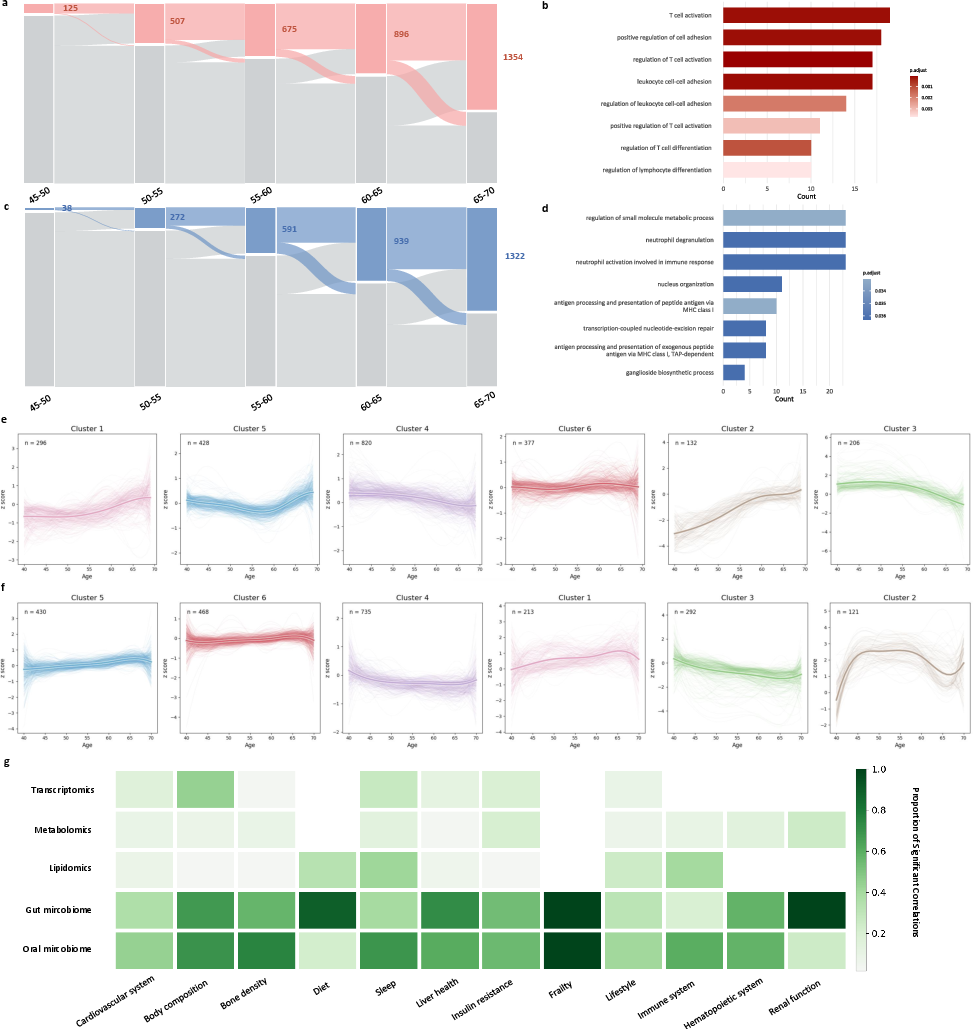
Figure 3: Sankey diagrams, pathway enrichment, and trajectory clusters reveal distinct molecular dynamics and systemic signatures in accelerated versus decelerated aging groups.
Pathway enrichment analyses highlight functional divergence: accelerated aging is enriched for T-cell activation and inflammatory pathways, while decelerated aging favors metabolic and redox homeostasis. Fuzzy C-means clustering of temporal trajectories reveals precipitous mid-life changes in accelerated aging, contrasted with gradual, late-life shifts in decelerated aging. The most pronounced group differences are concentrated in the gut and oral microbiomes, implicating a microbiome-centric axis in aging heterogeneity.
Temporal Waves and Sex-Specific Dynamics
Application of the DE-SWAN algorithm uncovers distinct temporal waves of molecular change across omics layers. Two consistent crests are observed: one around ages 45–55 and another near 60–65, followed by a decline after age 67. Transcriptomics and lipidomics show the largest amplitudes, while microbiome layers display concordant crests.
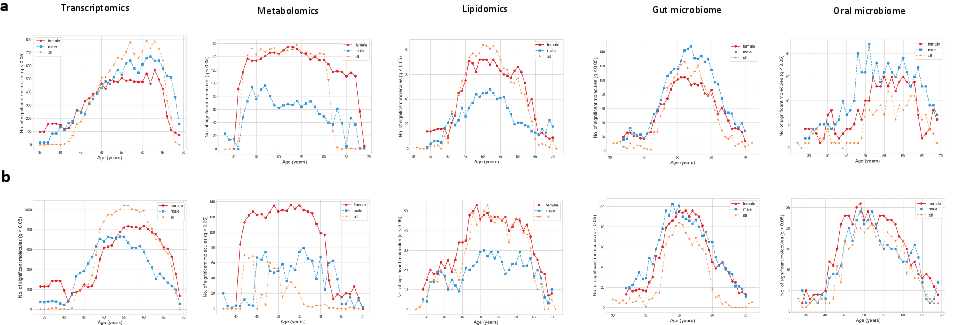
Figure 4: Temporal waves of age-associated molecular features in accelerated and decelerated aging, with sex-stratified analyses revealing distinct dynamics.
Sex-stratified analyses reveal higher crests in females for metabolomics and lipidomics, coinciding with the menopausal transition, while males exhibit delayed peaks in the oral microbiome and transcriptome. These findings suggest coordinated, system-level remodeling and highlight the importance of sex-specific factors in aging trajectories.
Clinical Relevance: Multimorbidity and Disease Associations
Regression analyses demonstrate significant correlations between multi-omics aging clocks and multimorbidity in older adults (>60 years), independent of chronological age, sex, and BMI. Microbiome AgeAccel scores show the strongest and broadest associations with 14 of 16 common non-cancer diseases, particularly metabolic disorders. Molecular omics AgeAccel scores derived from blood are more narrowly associated, primarily with cardiometabolic diseases.
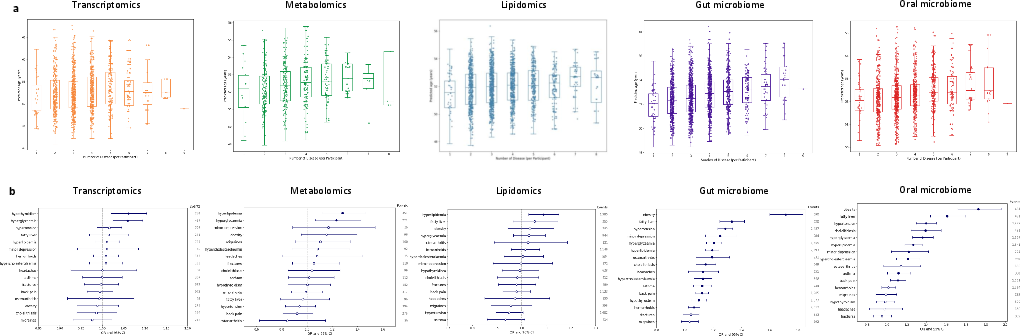
Figure 5: Multi-omics aging clocks are significantly associated with multimorbidity, mortality, and disease incidence, with microbiome clocks showing the strongest disease associations.
These results position multi-omics biological age as a promising surrogate endpoint for clinical trials and personalized risk stratification.
Methodological Considerations
The paper employs rigorous preprocessing pipelines for each omics layer, including quality control, normalization, and feature selection. Nonlinear modeling via LightGBM enables the capture of complex biological interactions and inflection points. The large, prospective cohort design and breadth of multi-omics profiling provide exceptional statistical power and generalizability, though the cohort’s ethnic homogeneity warrants caution in extrapolation to other populations.
Implications and Future Directions
This work reframes human aging as a dynamic, nonlinear, and heterogeneous process, with the microbiome emerging as a central indicator of healthspan. The identification of distinct aging archetypes and sex-specific molecular waves suggests critical windows for intervention, particularly in mid-life. The multi-omics aging clocks offer a robust framework for quantifying biological age and predicting disease risk, with potential applications in precision medicine, healthspan monitoring, and preventive strategies.
Future research should focus on validating these findings in diverse populations, leveraging longitudinal data to establish causality, and dissecting the mechanistic links between molecular changes and long-term health outcomes. The integration of additional omics layers (e.g., proteomics, epigenomics) and environmental exposures may further enhance the resolution and utility of aging clocks.
Conclusion
This paper establishes a comprehensive, multi-omics framework for quantifying biological age and decoding the molecular landscape of human aging. The integration of transcriptomic, metabolomic, lipidomic, and microbiome data enables the identification of distinct aging archetypes, sex-specific dynamics, and robust predictors of health outcomes. The findings underscore the centrality of the microbiome in aging and provide a foundation for personalized strategies to monitor healthspan and prevent age-related disease.




