- The paper introduces DrugReasoner, a reasoning-augmented LLM employing GRPO to provide interpretable drug approval predictions.
- The methodology integrates molecular descriptors with comparative reasoning against similar compounds to enhance prediction accuracy and transparency.
- Empirical results demonstrate robust performance with AUC scores around 0.73 and high recall, outperforming baselines including ChemAP on external datasets.
DrugReasoner: Interpretable Drug Approval Prediction with a Reasoning-Augmented LLM
Introduction
DrugReasoner introduces a reasoning-augmented LLM framework for interpretable drug approval prediction, leveraging the LLaMA-3.1-8B-Instruct architecture fine-tuned with group relative policy optimization (GRPO). The model is designed to address the dual challenge of predictive accuracy and interpretability in early-stage drug discovery, where the ability to rationalize predictions is critical for downstream decision-making. DrugReasoner integrates molecular descriptors and comparative reasoning against structurally similar compounds, outputting both a binary approval prediction and a stepwise rationale with a confidence score. This approach is positioned as an advancement over prior models such as ChemAP, which, despite strong performance, lack transparent reasoning capabilities.
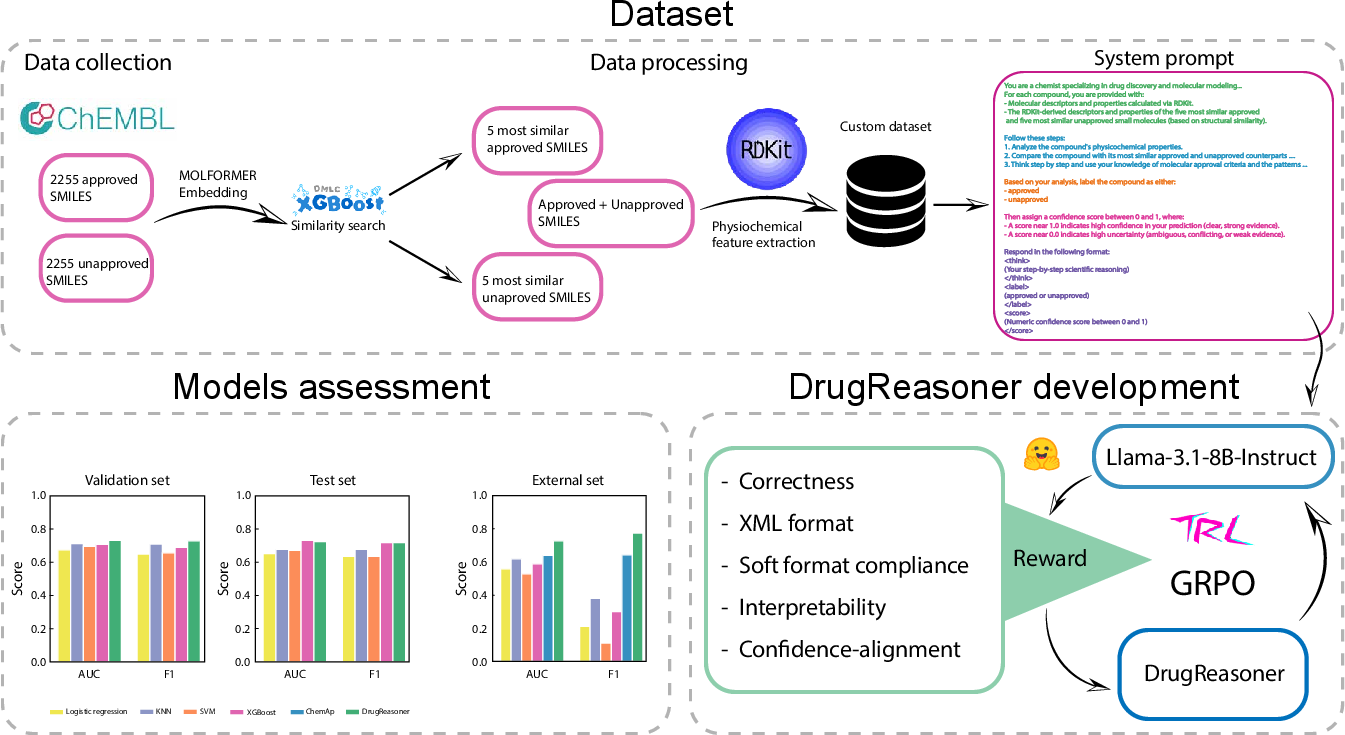
Figure 1: Schematic representation of DrugReasoner development and assessment, including dataset preparation, GRPO-based model training, and comparative evaluation against baselines.
Model Architecture and Training
Data Processing and Feature Engineering
DrugReasoner operates on a curated dataset of 2,255 approved and 2,255 unapproved small molecules from ChEMBL, with class balance achieved via random undersampling. Each molecule is represented by a set of physicochemical and structural descriptors computed using RDKit, explicitly excluding SMILES to mitigate data leakage from LLM pretraining. Embeddings are generated using MOLFORMER, a transformer-based model trained on SMILES, with mean pooling over the last hidden states to yield 768-dimensional representations.
For each query molecule, the five most similar approved and five most similar unapproved compounds are identified using XGBoost-based leaf embedding similarity, where Hamming distance in the decision space quantifies structural proximity. This comparative context is critical for the model's reasoning process.
Prompt Engineering and Output Schema
Inputs to DrugReasoner are structured prompts containing the RDKit descriptors of the query molecule and its ten nearest neighbors (five approved, five unapproved). The system prompt enforces a domain-specific instruction set to simulate expert chemical reasoning. The model outputs are formatted in XML with three fields: > (rationale), <label> (approved/unapproved), and <score> (confidence, 0.0–1.0), ensuring both interpretability and compatibility with reward modeling.
Fine-Tuning with Group Relative Policy Optimization
DrugReasoner is fine-tuned using GRPO, a reinforcement learning algorithm that extends PPO by leveraging group-based advantage estimation. For each input, the model generates Kj candidate responses, each scored by a multi-objective reward function. The group mean reward serves as a baseline, and the advantage for each response is computed as Ajk=Rjk−Rˉj. The surrogate loss incorporates a clipped objective and a KL-divergence penalty to stabilize updates.
The reward function is multi-faceted, combining correctness, XML format compliance, soft format compliance, interpretability, and confidence-alignment. This design ensures that the model not only predicts accurately but also adheres to the required output structure and provides semantically valid, confidence-calibrated rationales.
Training is performed for 14,500 steps on a single NVIDIA V100 GPU, with 4-bit quantization and LoRA applied to key attention projection layers to reduce memory and computational overhead. The optimal checkpoint is selected based on validation AUC and output structure adherence.
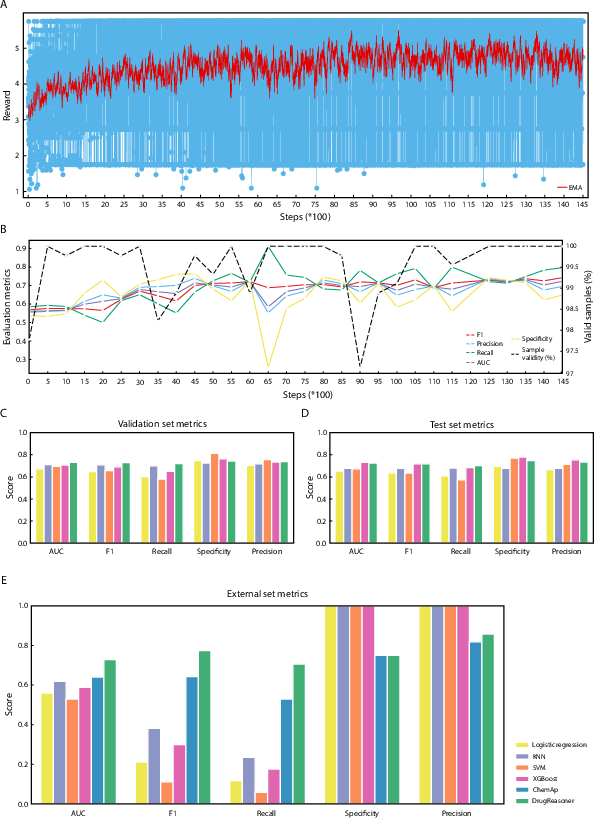
Figure 2: DrugReasoner assessment, including reward trajectory during GRPO training, evaluation metrics, and comparative performance against baselines and ChemAP on validation, test, and external datasets.
Empirical Results
On the validation set, DrugReasoner achieves an AUC of 0.732 and F1-score of 0.729, outperforming logistic regression, SVM, KNN, and matching or exceeding XGBoost. On the test set, the model maintains robust performance (AUC = 0.725, F1 = 0.718), with the highest recall (0.702) among all models. The model's confidence scores stabilize at 0.87, indicating reliable calibration.
External Dataset Generalization
On an independent external dataset (17 approved, 8 unapproved drugs), DrugReasoner achieves an AUC of 0.728 and F1-score of 0.774, outperforming all baselines and ChemAP (AUC = 0.64, F1 = 0.529). Notably, DrugReasoner attains the highest precision (0.857) and balanced accuracy (0.720), with strong recall (0.706) and specificity (0.750). Baseline models exhibit poor sensitivity (≤ 0.235) despite perfect specificity, underscoring DrugReasoner's superior generalizability and real-world applicability.
Interpretability and Output Structure
DrugReasoner consistently produces structured rationales that articulate the comparative reasoning process, referencing molecular features and similarities to known compounds. The model's output format is strictly enforced via reward shaping, resulting in 100% adherence to the expected XML schema at the selected checkpoint.
Implementation Considerations
Computational Requirements
Training DrugReasoner requires a single high-memory GPU (32 GB VRAM) and approximately 800 hours for full convergence. The use of 4-bit quantization and LoRA significantly reduces memory footprint, enabling efficient fine-tuning of the 8B-parameter LLaMA model. Inference can be performed on consumer-grade GPUs with reduced batch sizes.
Data Leakage Mitigation
To prevent memorization or data leakage from LLM pretraining, SMILES strings are excluded from model inputs. Only computed molecular descriptors are used, which also enhances interpretability. This design choice may limit the model's ability to capture fine-grained structural nuances but is justified by the need for robust generalization.
Reward Function Design
The multi-objective reward function is critical for aligning the model's outputs with both predictive accuracy and interpretability. The confidence-alignment component penalizes overconfident incorrect predictions and rewards calibrated uncertainty, improving trustworthiness in deployment.
Scaling and Future Extensions
The current implementation is constrained by context window (4,096 tokens) and model size (8B parameters). Scaling to larger models and longer contexts could enable the inclusion of richer structural information (e.g., SMILES for neighbors) and more complex reasoning chains. Systematic hyperparameter optimization and integration of additional domain knowledge (e.g., bioactivity, toxicity) are promising directions for further performance gains.
Theoretical and Practical Implications
DrugReasoner demonstrates that reasoning-augmented LLMs, when fine-tuned with structured comparative prompts and multi-objective RL, can achieve both high predictive accuracy and interpretable outputs in drug approval prediction. The model's ability to generalize to external datasets and provide transparent rationales addresses a key bottleneck in AI-assisted drug discovery, where trust and explainability are paramount.
The approach exemplifies a shift from black-box predictive models to transparent, reasoning-driven systems, aligning with regulatory and practical requirements in pharmaceutical R&D. The integration of GRPO enables efficient exploration of the output space, reinforcing desirable behaviors beyond simple accuracy.
Limitations and Future Directions
Key limitations include the exclusion of SMILES, which may restrict structural expressivity, and the computational cost of RL-based fine-tuning. The dataset size, while balanced, remains modest relative to the chemical space. Future work should explore controlled integration of structural data, scaling to larger models, and extension to multi-modal or multi-task settings (e.g., toxicity, efficacy, ADMET prediction). Incorporating active learning and uncertainty quantification could further enhance the model's utility in real-world pipelines.
Conclusion
DrugReasoner establishes a robust framework for interpretable drug approval prediction, combining LLM-based chain-of-thought reasoning, comparative molecular analysis, and reinforcement learning via GRPO. The model achieves strong empirical performance, outperforms established baselines and ChemAP on external data, and delivers structured, confidence-calibrated rationales. This work underscores the potential of reasoning-augmented LLMs as transparent, trustworthy tools for early-stage pharmaceutical decision-making and sets a foundation for future advances in interpretable AI for drug discovery.

