- The paper demonstrates that autonomous agent-based systems can match clinician-guided feature generation for predicting prostate cancer recurrence.
- The methodology employs specialized agents for feature discovery, extraction, validation, and aggregation to ensure high-quality, interpretable outputs.
- The study reveals that the AF system achieves an AUC-ROC comparable to manual methods, showcasing scalable clinical feature extraction.
Agent-Based Feature Generation from Clinical Notes for Outcome Prediction
Introduction
The paper "Agent-Based Feature Generation from Clinical Notes for Outcome Prediction" introduces a novel approach to extract meaningful features from unstructured clinical notes within Electronic Health Records (EHRs) using an autonomous system named \AFG. This system employs a modular multi-agent architecture powered by LLMs. It demonstrates that \AFG can generate structured clinical features autonomously and efficiently for predicting prostate cancer recurrence, matching the performance of manual Clinician Feature Generation (CFG) methods.
Methodology
The proposed \AFG pipeline utilizes specialized LLM agents, each responsible for distinct subtasks such as feature discovery, extraction, validation, post-processing, and aggregation. This agentic approach contrasts with existing methods that rely heavily on clinician guidance or simplistic representational models.
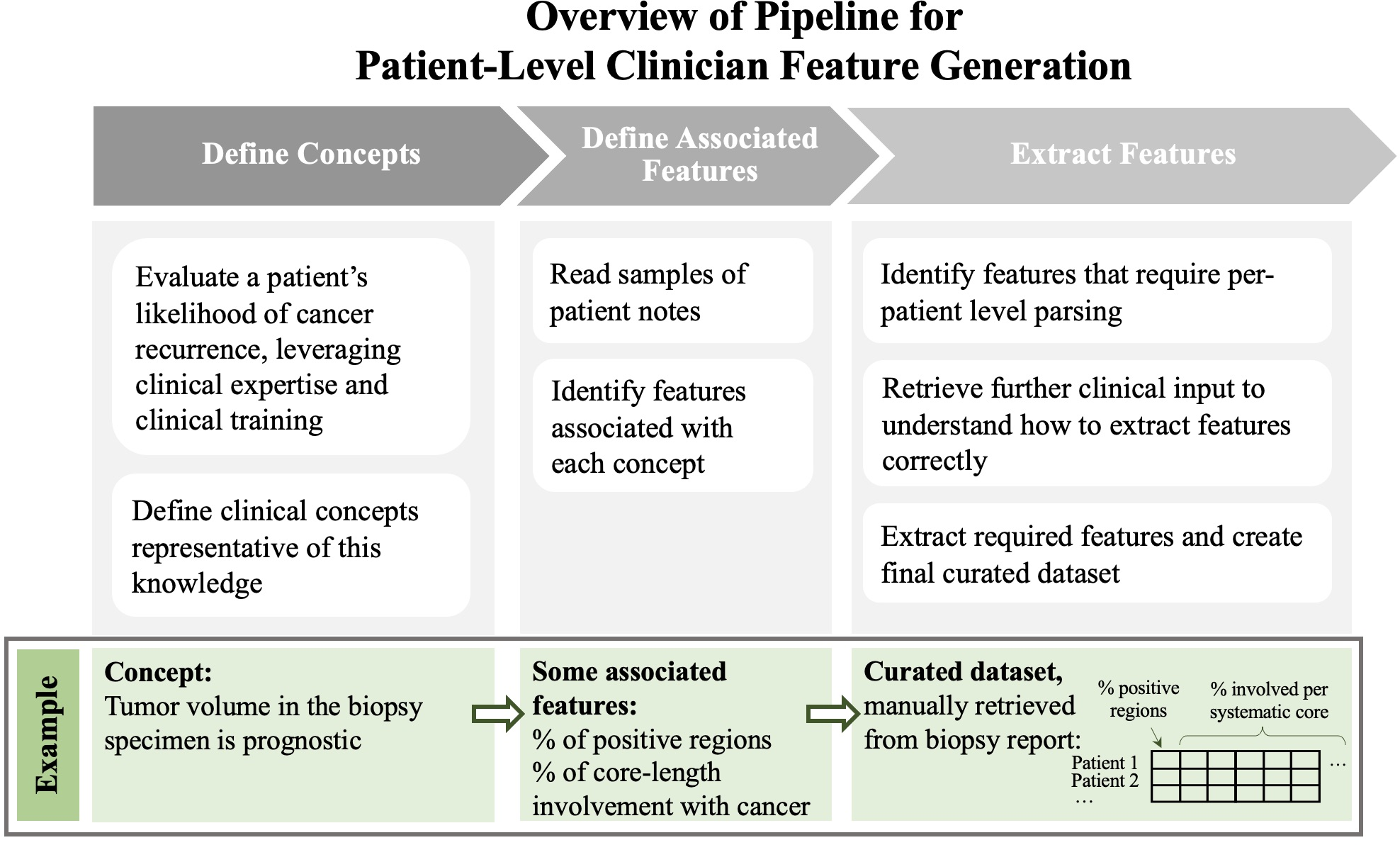
Figure 1: Overview of pipeline used to generate CFG features.
The specialized agents in \AFG work collaboratively:
- Feature Discovery Agent: Identifies potential structured variables from clinical notes, excluding features already present in structured datasets.
- Feature Extraction Agent: Retrieves values for proposed features from individual notes based on guidelines.
- Feature Validation Agent: Ensures quality control by assessing extracted values against a sample of notes, initiating revisions if necessary.
- Post-Processing and Aggregation Agents: Transform and aggregate features, employing Python code where required for complex calculations.
This approach allows \AFG to generate interpretable features autonomously, facilitating efficient integration into ML models.
Patient Cohort and Data
The paper evaluates \AFG on a cohort of 147 patients from Stanford Healthcare, focusing on predicting 5-year prostate cancer recurrence. Inclusion criteria involved specific prostate cancer treatments and data availability requirements. Through this paper, key patient-level features were extracted to assess the model's predictive capacity.
Results
The models were evaluated using nested cross-validation, comparing several feature generation techniques—including Baseline, CLFG, and CFG—alongside \AFG.
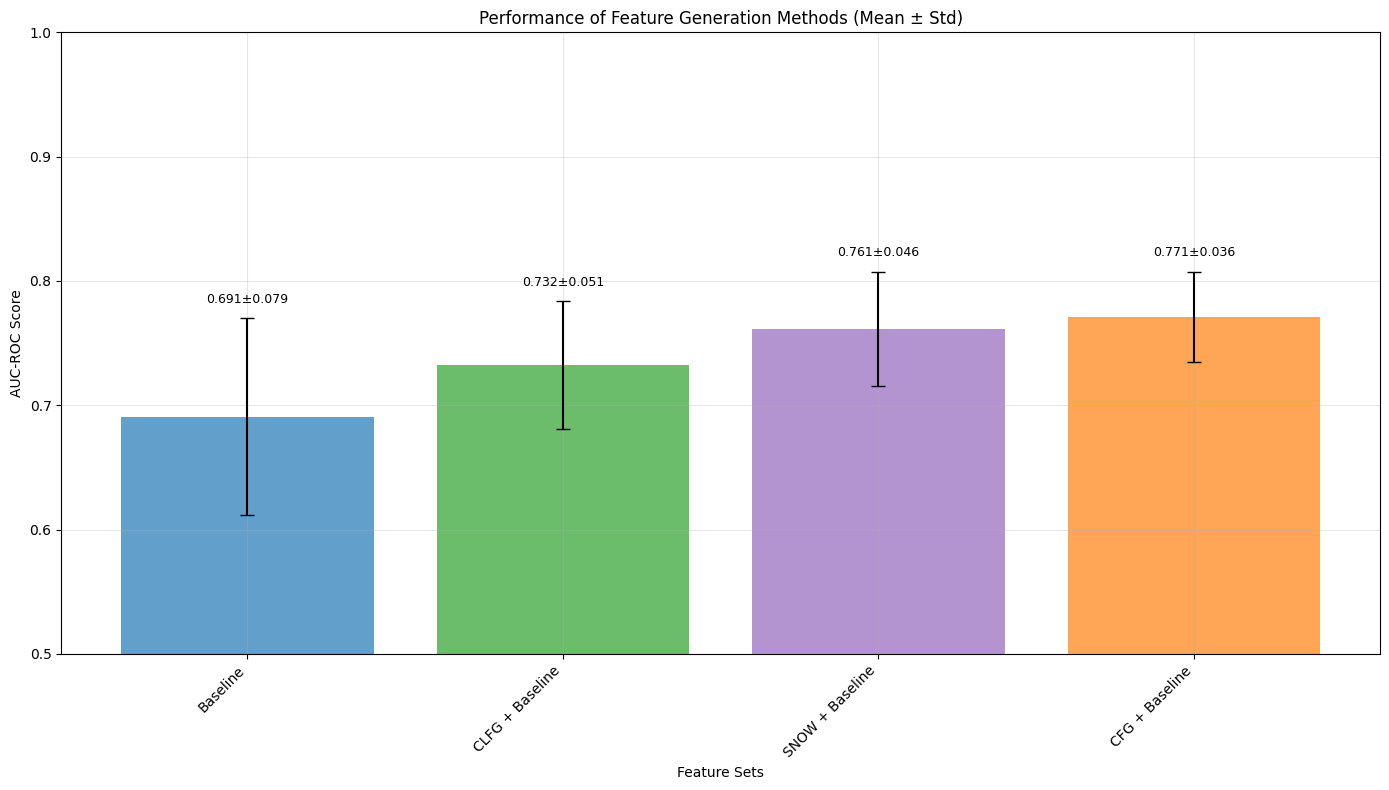
Figure 2: Comparison of the distribution of the AUC-ROC for different feature generation methods
- CFG Model: Achieved the highest mean AUC-ROC (0.771 ± 0.036), highlighting the significance of clinician expertise in feature generation.
- CLFG Model: Utilizing LLMs with clinician prompts reached an AUC-ROC of 0.732 ± 0.051, nearing manual CFG performance.
- \AFG Model: Matched CFG's performance with an AUC-ROC of 0.761 ± 0.046, confirming that autonomous systems can effectively replace manual processes.
Conversely, Representational Feature Generation (RFG) methods did not surpass baseline features in effectiveness, likely due to data dimensionality constraints.
Discussion
The \AFG system demonstrated equivalent performance to manual CFG processes, underscoring the potential of LLM agents to emulate expert-level feature engineering. By bridging the gap between manual extraction and scalable automation, \AFG provides an avenue to deploy structured, interpretable features in ML models without human intervention.
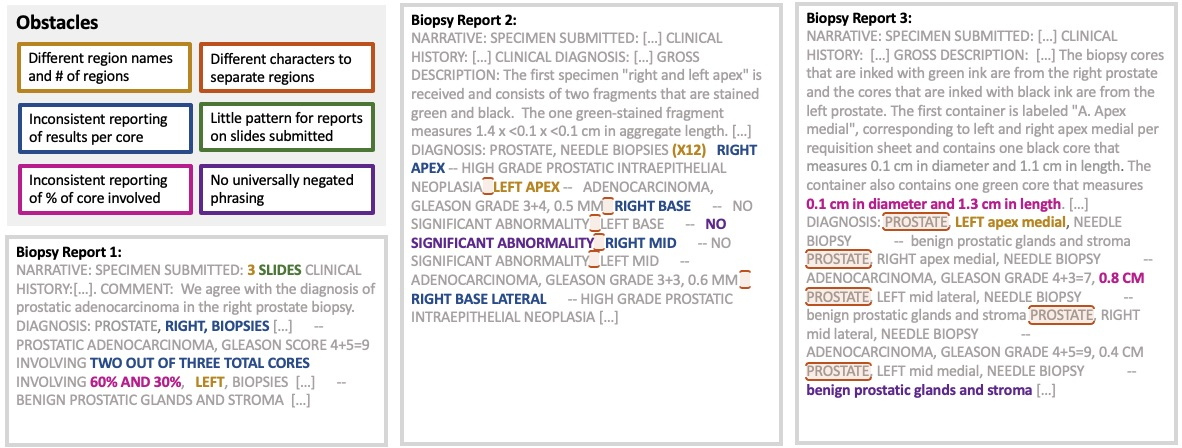
Figure 3: Examples of obstacles in automating the extraction of CFG features.
While RFG approaches show scalability, their lack of interpretability limits their applicability in clinical contexts. In contrast, \AFG maintains clinical relevance and detail—a critical component for high-stakes medical decision-making.
This research suggests a new paradigm for operationalizing clinical knowledge, where LLMs can autonomously generate and validate clinically meaningful features. As EHR data grows, systems like \AFG could offer scalable solutions for deploying AI-driven healthcare interventions.
Conclusion
The paper illustrates the viability of using agent-based architectures for extracting structured features from clinical notes. The \AFG system paves the way for scalable, interpretable, and clinically relevant feature generation, promising significant impact on ML implementations in health informatics. Future work may focus on broadening dataset sizes to leverage RFG methodologies effectively, further consolidating \AFG's place in clinical ML pipelines.


