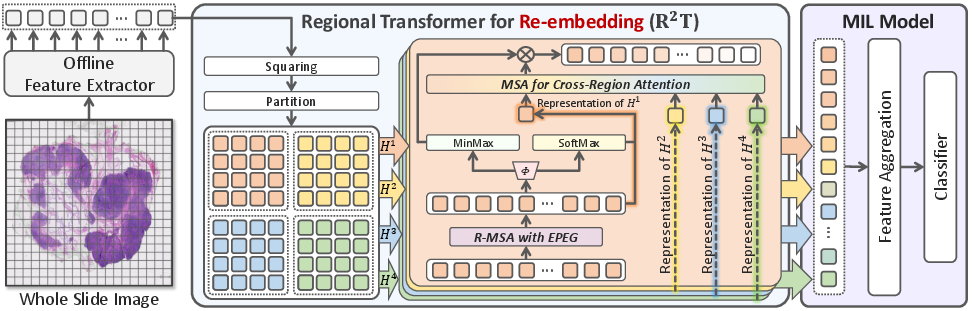
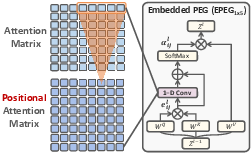
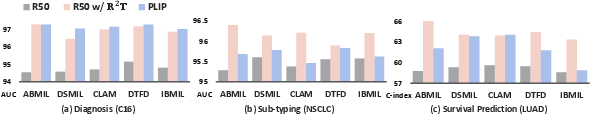
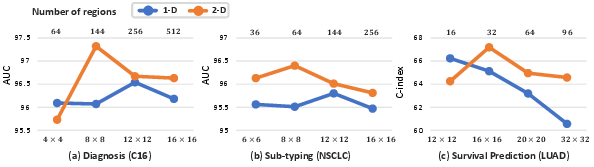
Feature Re-Embedding: Towards Foundation Model-Level Performance in Computational Pathology (2402.17228v4)
Abstract: Multiple instance learning (MIL) is the most widely used framework in computational pathology, encompassing sub-typing, diagnosis, prognosis, and more. However, the existing MIL paradigm typically requires an offline instance feature extractor, such as a pre-trained ResNet or a foundation model. This approach lacks the capability for feature fine-tuning within the specific downstream tasks, limiting its adaptability and performance. To address this issue, we propose a Re-embedded Regional Transformer (R$2$T) for re-embedding the instance features online, which captures fine-grained local features and establishes connections across different regions. Unlike existing works that focus on pre-training powerful feature extractor or designing sophisticated instance aggregator, R$2$T is tailored to re-embed instance features online. It serves as a portable module that can seamlessly integrate into mainstream MIL models. Extensive experimental results on common computational pathology tasks validate that: 1) feature re-embedding improves the performance of MIL models based on ResNet-50 features to the level of foundation model features, and further enhances the performance of foundation model features; 2) the R$2$T can introduce more significant performance improvements to various MIL models; 3) R$2$T-MIL, as an R$2$T-enhanced AB-MIL, outperforms other latest methods by a large margin.The code is available at: https://github.com/DearCaat/RRT-MIL.
- Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA, 318(22):2199–2210, 2017.
- Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nature Medicine, 25(8):1301–1309, 2019.
- Structure-aware transformer for graph representation learning. In International Conference on Machine Learning, pages 3469–3489. PMLR, 2022a.
- Scaling vision transformers to gigapixel images via hierarchical self-supervised learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 16144–16155, 2022b.
- Conditional positional encodings for vision transformers. arXiv preprint arXiv:2102.10882, 2021.
- Ai in computational pathology of cancer: Improving diagnostic workflows and clinical outcomes? Annual Review of Cancer Biology, 7:57–71, 2023.
- Artificial intelligence and computational pathology. Laboratory Investigation, 101(4):412–422, 2021.
- Imagenet: A large-scale hierarchical image database. In 2009 IEEE conference on computer vision and pattern recognition, pages 248–255. Ieee, 2009.
- An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929, 2020.
- Deep miml network. In Proceedings of the AAAI conference on artificial intelligence, 2017.
- Digital and computational pathology for biomarker discovery. Predictive Biomarkers in Oncology: Applications in Precision Medicine, pages 87–105, 2019.
- Deep residual learning for image recognition. In CVPR, pages 770–778, 2016.
- A visual–language foundation model for pathology image analysis using medical twitter. Nature Medicine, pages 1–10, 2023.
- Attention-based deep multiple instance learning. In ICML, pages 2127–2136. PMLR, 2018.
- Weakly-supervised learning for lung carcinoma classification using deep learning. Scientific reports, 10(1):9297, 2020.
- Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980, 2014.
- Albert: A lite bert for self-supervised learning of language representations. arXiv preprint arXiv:1909.11942, 2019.
- Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning. In CVPR, pages 14318–14328, 2021a.
- Dt-mil: Deformable transformer for multi-instance learning on histopathological image. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 206–216. Springer, 2021b.
- Interventional bag multi-instance learning on whole-slide pathological images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 19830–19839, 2023.
- Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, pages 10012–10022, 2021.
- Data-efficient and weakly supervised computational pathology on whole-slide images. Nature Biomedical Engineering, 5(6):555–570, 2021a.
- Data-efficient and weakly supervised computational pathology on whole-slide images. Nature biomedical engineering, 5(6):555–570, 2021b.
- Learning transferable visual models from natural language supervision. In International conference on machine learning, pages 8748–8763. PMLR, 2021.
- Self supervised learning improves dmmr/msi detection from histology slides across multiple cancers. arXiv preprint arXiv:2109.05819, 2021.
- Transmil: Transformer based correlated multiple instance learning for whole slide image classification. NeurIPS, 34, 2021.
- Cluster-to-conquer: A framework for end-to-end multi-instance learning for whole slide image classification. arXiv preprint arXiv:2103.10626, 2021.
- Self-attention with relative position representations. arXiv preprint arXiv:1803.02155, 2018.
- Artificial intelligence for digital and computational pathology. Nature Reviews Bioengineering, pages 1–20, 2023.
- Multiple instance learning framework with masked hard instance mining for whole slide image classification. In Proceedings of the IEEE/CVF International Conference on Computer Vision, pages 4078–4087, 2023.
- Personalized breast cancer treatments using artificial intelligence in radiomics and pathomics. Journal of medical imaging and radiation sciences, 50(4):S32–S41, 2019.
- Laurens Van der Maaten and Geoffrey Hinton. Visualizing data using t-sne. Journal of machine learning research, 9(11), 2008.
- Attention is all you need. Advances in neural information processing systems, 30, 2017.
- Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE transactions on cybernetics, 50(9):3950–3962, 2019.
- Lymph node metastasis prediction from whole slide images with transformer-guided multi-instance learning and knowledge transfer. IEEE Transactions on Medical Imaging, 2022.
- Deep learning in digital pathology for personalized treatment plans of cancer patients. In Seminars in Diagnostic Pathology, pages 109–119. Elsevier, 2023.
- Rethinking and improving relative position encoding for vision transformer. In Proceedings of the IEEE/CVF International Conference on Computer Vision, pages 10033–10041, 2021.
- Deep learning-based survival prediction for multiple cancer types using histopathology images. PloS one, 15(6):e0233678, 2020.
- Evaluating cancer-related biomarkers based on pathological images: a systematic review. Frontiers in Oncology, 11:763527, 2021.
- Nyströmformer: A nyström-based algorithm for approximating self-attention. In Proceedings of the AAAI Conference on Artificial Intelligence, pages 14138–14148, 2021.
- Camel: A weakly supervised learning framework for histopathology image segmentation. In CVPR, pages 10682–10691, 2019.
- Graphformers: Gnn-nested transformers for representation learning on textual graph. Advances in Neural Information Processing Systems, 34:28798–28810, 2021.
- Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks. Medical Image Analysis, 65:101789, 2020.
- A transformer-based framework for multivariate time series representation learning. In Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining, pages 2114–2124, 2021.
- Dtfd-mil: Double-tier feature distillation multiple instance learning for histopathology whole slide image classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 18802–18812, 2022a.
- Dual space multiple instance representative learning for medical image classification. In 33rd British Machine Vision Conference 2022, BMVC 2022, London, UK, November 21-24, 2022. BMVA Press, 2022b.
- Predicting lymph node metastasis using histopathological images based on multiple instance learning with deep graph convolution. In CVPR, pages 4837–4846, 2020.
- Setmil: spatial encoding transformer-based multiple instance learning for pathological image analysis. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2022: 25th International Conference, Singapore, September 18–22, 2022, Proceedings, Part II, pages 66–76. Springer, 2022.
- Cross-modal translation and alignment for survival analysis. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), pages 21485–21494, 2023.
- Wsisa: Making survival prediction from whole slide histopathological images. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 7234–7242, 2017.
Paper Prompts
Sign up for free to create and run prompts on this paper using GPT-5.
Top Community Prompts
Collections
Sign up for free to add this paper to one or more collections.