- The paper introduces RADiAnce, a framework that integrates retrieval and conditional latent diffusion to enhance cross-domain protein binder design.
- The method employs an all-atom VAE with contrastive learning and dot-product similarity to accurately retrieve interface templates for binder generation.
- The framework outperforms baselines by achieving high recall and precision, enabling de novo antibody design without predefined structures.
Latent Retrieval Augmented Generation of Cross-Domain Protein Binders
Introduction
The design of protein binders, which target specific binding sites on proteins, is a vital aspect of drug discovery and structural biology. This paper introduces "Latent Retrieval Augmented Generation of Cross-Domain Protein Binders," focusing on the Retrieval-Augmented Diffusion framework for Aligned interface (RADiAnce) to enhance the generation of novel binders by leveraging existing binding interfaces. Traditional generative models struggle to rationally and interpretably generate effective binders. RADiAnce innovatively unifies retrieval and generation processes in a shared contrastive latent space, allowing for efficient retrieval of relevant interfaces and facilitating their integration through a conditional latent diffusion generator.

Figure 1: Visualization of interface similarity across antibodies, proteins, and peptides, highlighting similar interaction patterns among diverse binder types.
Methodology
Contrastive Latent Space
RADiAnce relies on an all-atom variational autoencoder (VAE) augmented with contrastive learning to create an interaction-aligned latent space. This space allows the mapping of binding sites and interfaces to latent vectors that align for positive pairs while repelling negative ones. The retrieval of interfaces uses a dot-product similarity in this latent space, enabling fast and accurate identification of structurally relevant examples. Subsequently, the diffusion model operates in the same latent space, utilizing retrieved interface embeddings to condition binder generation.

Figure 2: Overview of RADiAnce framework, illustrating cross-domain encoding, alignment via contrastive loss, and the conditional diffusion generator's iterative refinement process.
Retrieval-Conditioned Latent Diffusion
RADiAnce introduces a retrieval-conditioned latent diffusion model to facilitate sequence-structure codesign. The diffusion process transforms latent variables through controlled noise addition and removal, leveraging retrieved samples as contextual templates during denoising. Prompt integration through cross-attention mechanisms ensures that the model effectively incorporates relevant interaction motifs into the generated binders.
Experiments
Retrieval Reliability
The retrieval component of RADiAnce demonstrates robust performance, achieving high recall and precision across antibody and peptide datasets. Critical findings suggest that exposure to diverse interface types significantly enhances retrieval efficacy, underscoring the importance of cross-domain information for binder generation.
RADiAnce outperforms several strong baseline models across established structural and biochemical metrics in both peptide and antibody binder generation tasks. Enhanced accuracy in recovering sequences, structures, and interaction patterns showcase the framework's ability to leverage retrieved interface knowledge efficiently.
Antibody Design Without Predefined Structures
In practical applications, RADiAnce supports de novo antibody design without the need for predefined structural frameworks. Iterative redesign cycles guided by retrieval enhance binding affinity and specificity, demonstrating the framework's potential for real-world antibody design.

Figure 3: Examples of de novo antibody designs targeting the HIV-1 receptor CD4, illustrating the successful interaction designs guided by RADiAnce.
Detailed Analysis
The study explores the impact of retrieval strategy on generative performance, affirming that adaptive retrieval aids superior sequence and structural generation by dynamically filtering out less relevant templates. Cross-domain retrieval boosts model effectiveness, establishing RADiAnce as a paradigm for unified retrieval-augmented generative frameworks.
Conclusion
RADiAnce effectively bridges the gap between retrieval-based knowledge incorporation and generative modeling, enabling rational and interpretable design of protein binders across domains. The method's performance depends heavily on retrieval quality, suggesting future enhancements in structural descriptors could further improve integration strategies. Ultimately, RADiAnce offers a transparent, controllable binder design pathway, with promise for accelerating therapeutic molecule development.
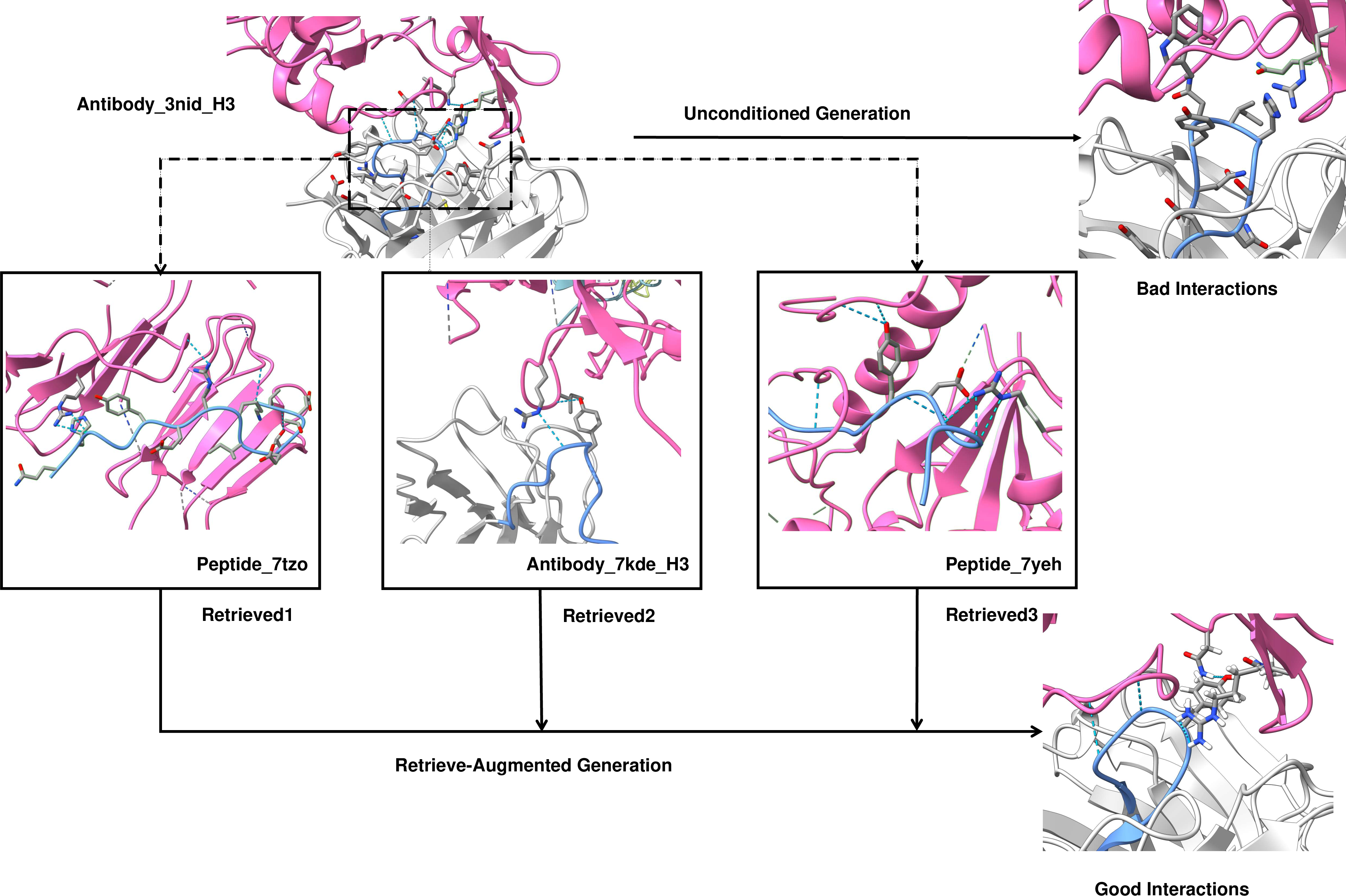
Figure 4: Case study of HCDR3 design for the GPIIb/IIIa binder showing the preservation of key interaction modes through retrieval-guided generation.





