- The paper presents AbMEGD, a novel framework that employs multi-scale equivariant graph diffusion to co-design antibody sequences and structures for precise antigen binding.
- It leverages advanced geometric modeling—integrating atomic and residue-level features—to outperform existing methods like DiffAb, especially in the CDR-H3 region.
- The study demonstrates that applying forward and backward diffusion processes optimizes antibody structures, enhancing binding affinity and structural integrity.
Antibody Design and Optimization with Multi-scale Equivariant Graph Diffusion Models for Accurate Complex Antigen Binding
Introduction
The paper presents AbMEGD, a novel computational framework addressing the challenges of antibody design for complex antigens. Traditional methods face limitations in accurately modeling geometric features while preserving symmetries and generalizing to novel antigen interfaces. AbMEGD leverages geometric deep learning, specifically multi-scale equivariant graph diffusion, to achieve precise antibody sequence and structure co-design. The approach integrates atomic-level geometric features with residue-level embeddings, providing a comprehensive solution to the geometric and generalization challenges inherent in antibody design.
Methodology
AbMEGD employs a multi-scale architecture incorporating multi-level representations to capture both global and atomic-level interactions crucial for antibody-antigen binding. The framework utilizes advanced geometric operations, including bond length, bond angle, and torsion calculations, akin to classical molecular dynamics, ensuring precise geometric modeling:

Figure 1: The antibody-antigen complex structure and CDRs involved in AbMEGD's geometric calculations. This figure translates bonded terms from classical molecular dynamics (MD)—such as bond length, bond angle, dihedral torsion, and improper angle—into efficient linear-time model operations.
The generative diffusion process is pivotal to AbMEGD, where the current state of CDRs guides the modeling of subsequent distributions for sequences and orientations:

Figure 2: This figure depicts the generative diffusion process. At every iteration, the network uses the current state of the CDR as input to model the distribution for the sequences, positions, and orientations of the CDR in the subsequent step.
Sequence-Structure Co-design
AbMEGD demonstrates robust performance across key metrics, outperforming leading models like DiffAb in sequence-structure co-design, notably in the critical CDR-H3 region. The results indicate significant improvements in amino acid recovery and structural accuracy:

Figure 3: Examples of CDR-H3 designed using a sequence-structure co-design approach, showcasing the distributions of interaction energy (ΔG) and RMSD. The antigen-antibody complex is derived from PDB entry, with the antigen being the SARS-CoV-2 RBD. Sample 1 exhibits better complementarity with the antigen, while Sample 3 displays less effective binding, potentially explaining the variation in their interaction energy (ΔG).
Antibody Optimization
AbMEGD's optimization capability further enhances antibody design by refining existing structures to improve binding affinity. The framework efficiently maintains structural integrity while achieving energy improvements across various optimization steps:
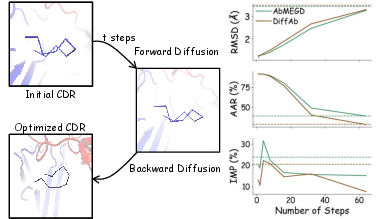
Figure 4: (a) The antibody fenoptimization algorithm applies a forward diffusion process to perturb the initial CDR over t steps, followed by a backward diffusion process to refine it into the optimized CDR. (b) The IMP, RMSD, and SeqID metrics of the CDRs were evaluated across varying optimization steps. The dashed lines indicate the performance of CDRs generated through de novo design. At t=4, the optimized CDRs achieve an IMP score comparable to that of the de novo designs while retaining a structural similarity to the original CDR.
Conclusion
AbMEGD presents a sophisticated approach to antibody design, demonstrating significant advancements in accuracy and efficiency through a unified framework. The integration of multi-scale equivariant graph diffusion enhances both sequence co-design and optimization tasks, establishing a benchmark for future therapeutic antibody development. As computational techniques evolve, integrating models like AbMEGD with broader datasets and more complex antigen interfaces could drive further innovations in computational immunology and therapeutic design.



