- The paper demonstrates that a shared encoder efficiently integrates diverse data by leveraging parameter sharing and modality-specific tokens.
- The paper introduces a methodology using transformer layers with appended modality features to enhance performance in data-scarce medical environments.
- The paper shows significant empirical gains on datasets like PMC-OA and MIMIC-CXR, indicating improved generalization over modality-specific encoders.
A Shared Encoder Approach to Multimodal Representation Learning
Introduction and Motivation
Multimodal representation learning stands out as a critical technique in machine learning, enabling models to integrate diverse data types, such as text, images, audio, and video, into cohesive representations. This capability is particularly transformative in domains like medicine, where heterogeneous data sources like radiology images, clinical notes, and EHR can be leveraged to improve diagnostic accuracy and patient care. However, medical applications face significant obstacles due to data scarcity, privacy concerns, and the labor-intensive nature of data labeling, which hinders the effective training of modality-specific encoders.
The paper "A Shared Encoder Approach to Multimodal Representation Learning" (2503.01654) introduces a novel shared encoder framework designed to address these challenges. By employing a single encoder across all modalities, the approach promises efficient representation learning, especially in low-data environments. This strategy not only curtails the risk of overfitting but also promotes enhanced generalization by capitalizing on shared structures within medical data.
Methodological Innovations
The shared encoder framework posited by this paper operates on the principle of parameter sharing across different modalities. This core innovation is augmented by learnable modality features that encode modality-specific information, allowing the shared encoder to differentiate inputs from various sources. The architecture simplifies adaptation to new modalities and tasks with limited labeled data, making it highly applicable to real-world medical scenarios.
Modality Identifier and Shared Encoder
The proposed system introduces two methods to incorporate modality-specific information: appending a modality feature vector to input embeddings, or attaching a modality token at the sequence's start. These strategies bolster the encoder's ability to discern between modalities without losing the homogenized structure essential for multimodal synergy.
The shared encoder itself is constructed from a sequence of transformer layers, which are universally applicable across all input types. This uniform application ensures computational efficiency while facilitating robust cross-modal learning. Early and late modality-specific layers can be integrated to capture essential low-level features or provide a nuanced balance between generalization and specialization.
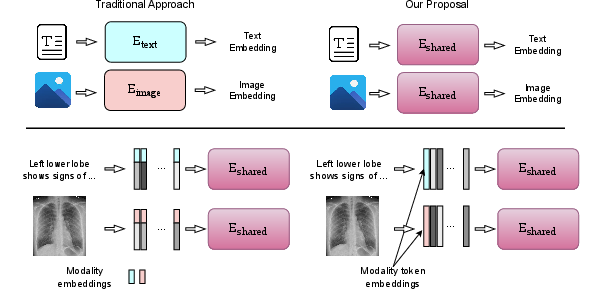
Figure 1: Top: Encoding data with (left) individual encoders vs (right) a shared encoder. Bottom: Adding modality-specific information by (left) appending modality embeddings to feature vectors, vs (right) adding a modality token.
Empirical Evaluation
The empirical results underscore the effectiveness of the shared encoder framework in multimodal representation learning compared to traditional modality-specific models. Evaluations conducted on the PMC-OA dataset for pretraining, as well as retrieval tasks involving MIMIC-CXR, Quilt, and DeepEyeNet datasets, reveal that the shared encoder outperforms disjoint modality-specific encoders in low-data settings. The introduction of modality-specific embeddings or specialized layers further enhances performance.
Impact on Low-Data Regimes
A key insight from the experiments is the shared encoder's pronounced advantage in scenarios with limited training data. Performance comparisons across different data sizes demonstrate notable gains using the shared approach, reaffirming its capacity to generalize effectively without exhaustive training data. Table summaries highlight relative improvements in retrieval tasks, reinforcing the model's adaptability and efficiency.
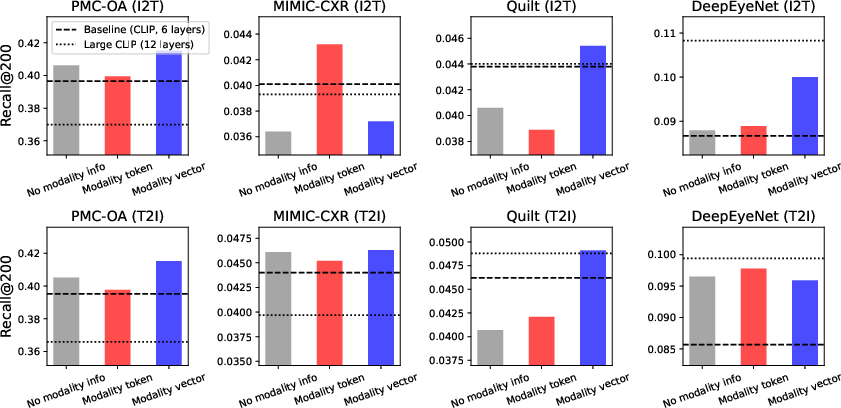
Figure 2: Comparison of methods for incorporating modality information into a shared encoder. The baseline (dark dashed line) uses two disjoint modality-specific encoders (125M parameters total), while the Large CLIP model (dotted line) has 210M parameters. All shared encoders have 125M parameters.
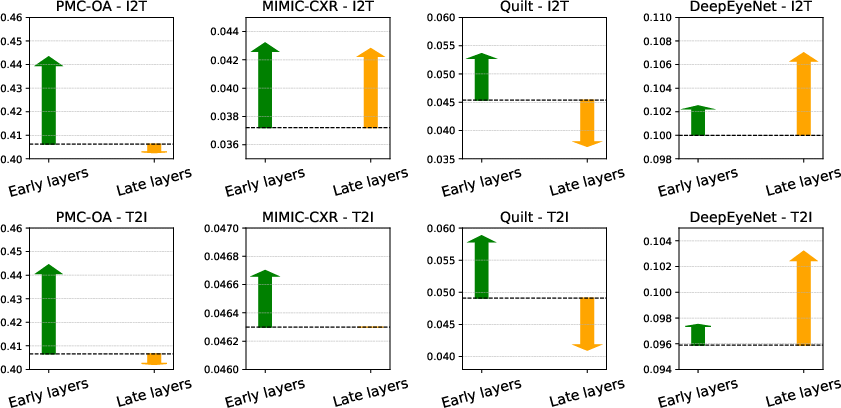
Figure 3: Impact of adding modality-specific layers to encoders. The dashed line shows the performance of a purely shared encoder, with green arrows indicating performance gains from early modality layers and orange arrows showing gains (or losses) from late modality layers.
Conclusion
The paper presents compelling evidence for the proposed shared encoder framework's potential to revolutionize multimodal representation learning, especially in the medically-focused datasets where the scarcity of labeled samples is a significant barrier. By leveraging shared parameters and strategic modality-specific enhancements, the framework ensures robust performance under data constraints. The anticipated release of implementation resources will enable further research, potentially catalyzing advancements in understanding and applying multimodal learning in complex domains such as medicine.




