- The paper presents a methodology leveraging LLMs to semi-automate clinical trial ontology creation, reducing reliance on manual data processing.
- It compares automated methods with human processes using cost, time, and coverage metrics, showing notable improvements in efficiency.
- Evaluation using OQuaRE metrics highlights both the promise and limitations, suggesting future work on relationship mapping and hallucination reduction.
Clinical Trials Ontology Engineering with LLMs
The paper "Clinical Trials Ontology Engineering with LLMs" explores the application of LLMs such as GPT3.5, GPT4, and Llama3 in managing clinical trial information by automating the extraction and integration of data into ontologies. This approach addresses the inefficiencies and high costs associated with manual processing of clinical trial data. This essay examines the methodology and findings of the paper, focusing on the operational aspects, evaluation metrics, and implications for future AI developments in medical research.
Methodology
The proposed methodology centers on leveraging LLMs to semi-automate the conversion of clinical trial data into ontological frameworks, facilitating real-time data integration. This involves the creation of ontologies based on clinical trials data sourced from clinicaltrials.gov, focusing on diseases like diabetes.
The process initiates with the extraction of relevant data using LLMs, followed by ontology creation and merging. Each LLM-generated ontology is integrated into a single main ontology through a novel merging method designed specifically for handling outcomes from clinical trials. Its scalability stems from organizing data at the triple level and employing a sorted synonym list to minimize duplication, thereby maintaining operational efficiency.
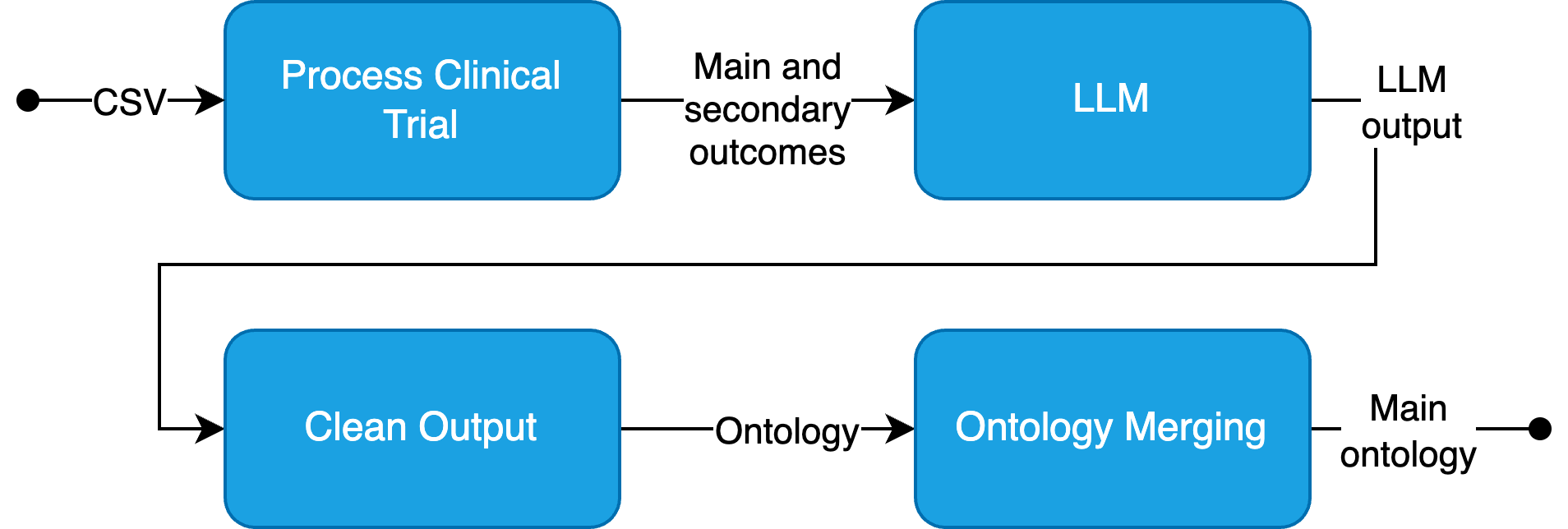
Figure 1: High-overview of proposed methodology.
Evaluation
Practical Metrics
The paper evaluates models based on cost, time, and inclusivity of generated ontologies. Cost analysis considers both LLM operation costs and human processing extrapolated from limited samples. Time metrics involve the duration taken per trial and overall generation times across models.
| Model |
Total Cost / Trial |
Total Time / Trial |
Included Ontologies |
| Human |
≈ \$5 |≈ 15 min. |
100\% |
|
| GPT3 |
\$0.0054 |
143 sec. |
76\% |
| chainedGPT3 |
\$0.0072 |
210 sec. |
80\% |
| GPT4 |
\$0.0624 |
107 sec. |
26\% |
| chainedGPT4 |
\$0.0941 |
212 sec. |
86\% |
| Llama3 (8b) |
\$0.0053 |
56 sec. |
28\% |
| chainedLlama3 (8b) |
\$0.0082 |
87 sec. |
24\% |
| Llama3 (70b) |
\$0.0579 |
110 sec. |
54\% |
| chainedLlama3 (70b) |
\$0.0898 |
171 sec. |
74\% |
OQuaRE Metrics
OQuaRE framework evaluates quality metrics like NOCOnto, highlighting extraction efficiency across models. Chained models generally outperform non-chained ones, with GPT4 displaying substantial improvements when prompting techniques are applied.
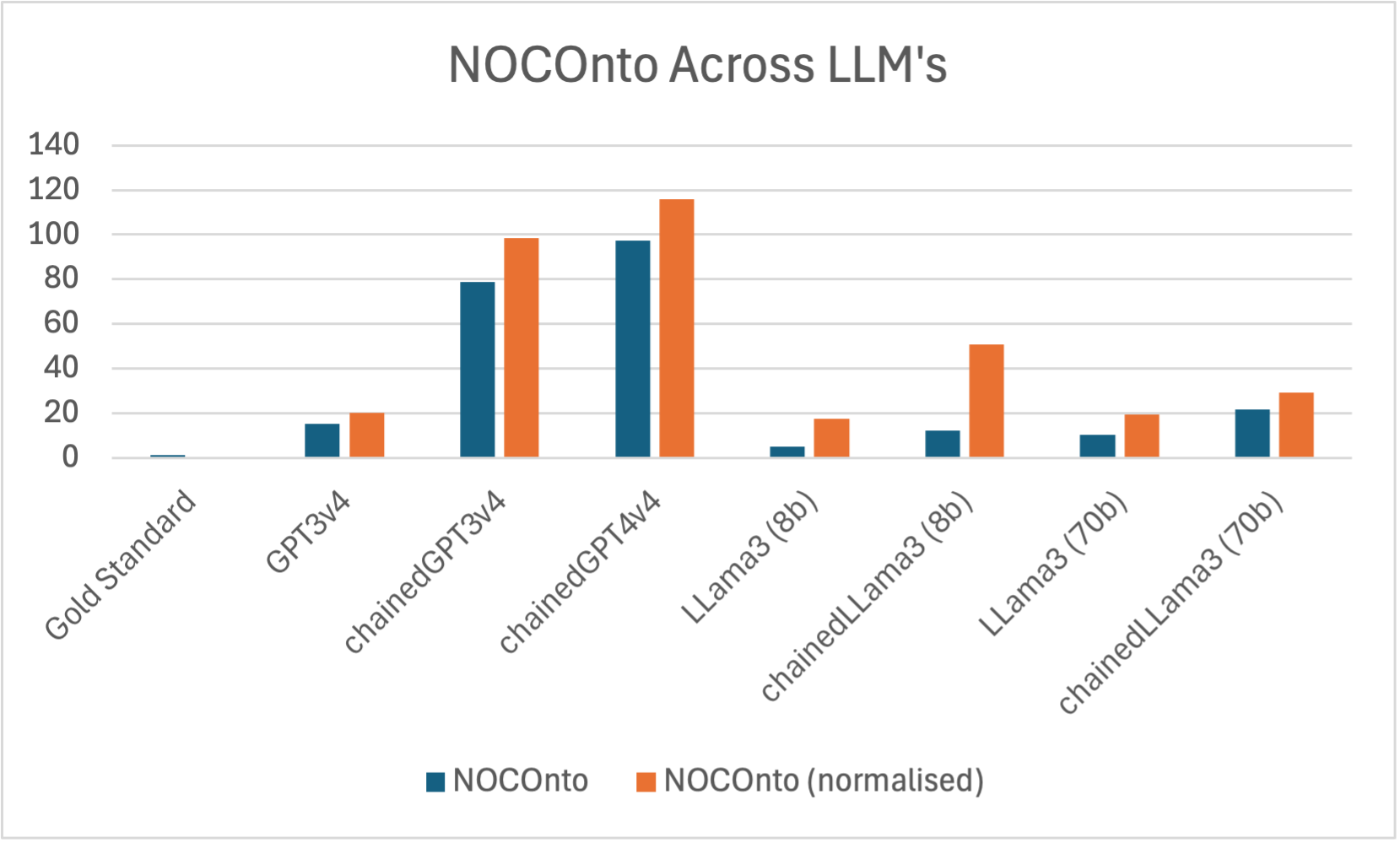
Figure 2: NOCOnto metric across different LLMs and used techniques.
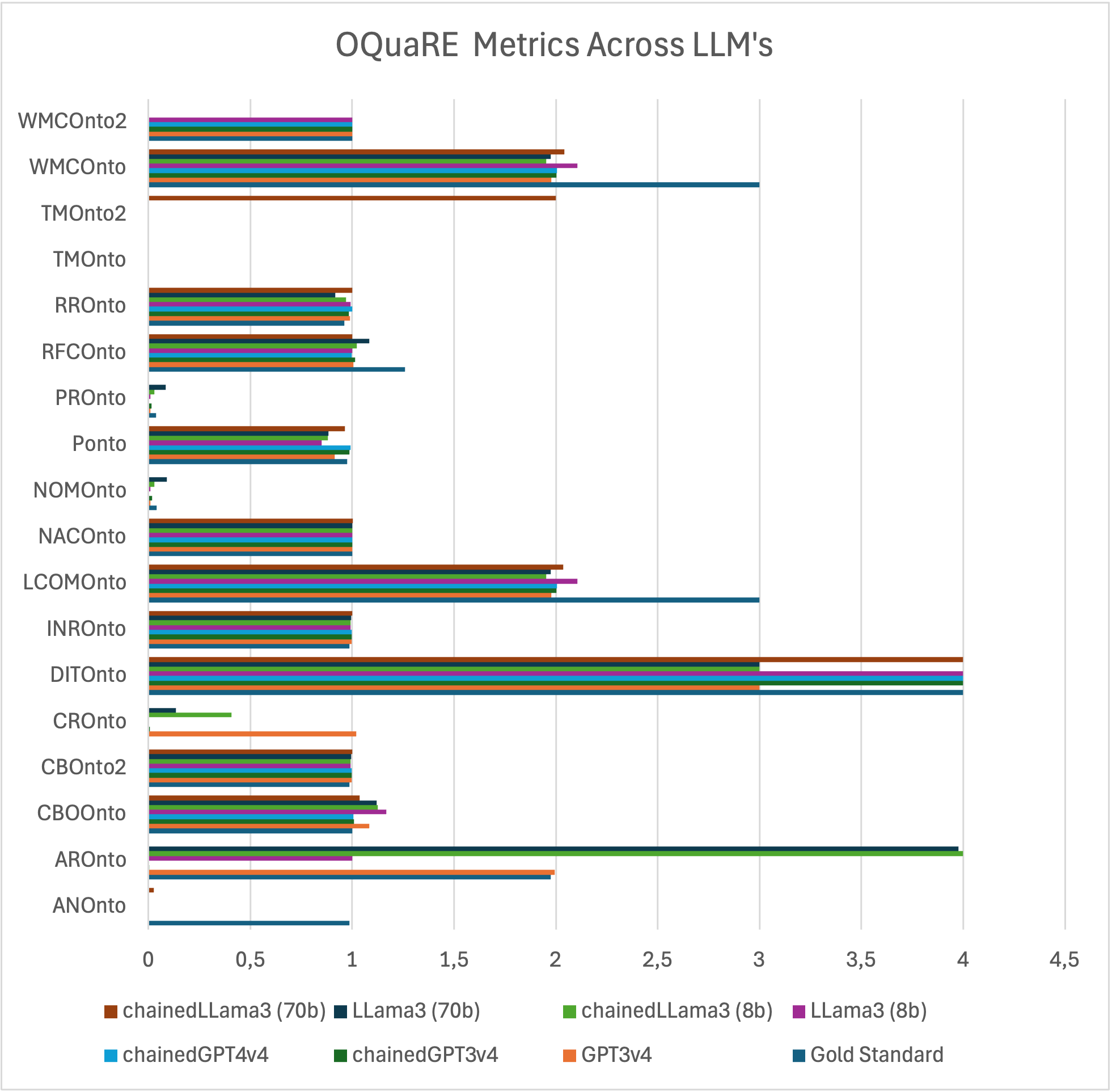
Figure 3: All OQuaRE metrics across different LLMs and techniques.
Discussion
Implications
The paper suggests that deploying LLMs for clinical trial ontology engineering is feasible both economically and temporally. These models significantly reduce processing costs compared to human efforts, suggesting potential shifts in how medical data management might be operationalized for real-time access.
Limitations
While LLMs exhibit efficiency, limitations revolve around syntax validity and the lack of relationship preservation between concepts. Addressing hallucinations and developing more robust ontology merging protocols to preserve inter-concept relationships are crucial areas for future research.
Conclusion
The integration of LLMs into clinical trials data processing showcases a pragmatic step towards AI-assisted medical data management. By lowering costs and accelerating processing times, models like GPT4, especially when enhanced through strategic prompting, hold promise for transforming large-scale data integration practices in clinical settings. Future work remains necessary to enhance relationship mapping within ontologies and further mitigate data hallucinations.




