- The paper presents SkinGenBench as a benchmark assessing how preprocessing pipelines and generative models affect synthetic dermoscopic image fidelity and diagnostic performance.
- The study compares StyleGAN2-ADA and DDPMs, showing that GAN-generated images improve class coherence and boost melanoma detection metrics by up to 15% F1-score.
- The analysis indicates that while advanced preprocessing marginally refines image quality, preserving clinical texture cues is crucial for reliable diagnostics.
SkinGenBench: Generative Model and Preprocessing Effects for Synthetic Dermoscopic Augmentation in Melanoma Diagnosis
Introduction
The study titled "SkinGenBench: Generative Model and Preprocessing Effects for Synthetic Dermoscopic Augmentation in Melanoma Diagnosis" (2512.17585) introduces SkinGenBench, a comprehensive benchmark designed to evaluate the interaction between preprocessing complexity and generative model choice in the context of synthetic dermoscopic image augmentation and melanoma diagnosis. Melanoma remains a critical public health challenge, with early detection significantly improving survival rates. The paper leverages a dataset comprised of 14,116 dermoscopic images from HAM10000 and MILK10K, encompassing five types of skin lesions: Nevus (NV), Basal Cell Carcinoma (BCC), Benign Keratosis Like (BKL), Melanoma (MEL), and Squamous Cell Carcinoma (SCC) (Figure 1).
The study evaluates two prominent generative paradigms, StyleGAN2-ADA and Denoising Diffusion Probabilistic Models (DDPMs), under varying preprocessing conditions. The focus of this research is to understand how preprocessing and generative model choices influence image quality and downstream diagnostic performance.

Figure 1: The types of skin lesions used in this study and their distribution in the curated dataset. Namely Nevus (NV, 52.60\%), Basal Cell Carcinoma (BCC, 21.43\%), Benign Keratosis Like (BKL, 11.60\%), Melanoma (MEL, 11.03\%), and Squamous Cell Carcinoma (SCC, 3.34\%).
Methodology
The study's experimental design is depicted in Figure 2. It begins with two preprocessing pipelines, basic and advanced, applied to the curated dataset. The basic pipeline involves geometric augmentations such as rotations and flips, while the advanced pipeline incorporates artifact removal techniques aimed at eliminating common artifacts like hair and ruler marks using the Dullrazor algorithm (Figure 3). The objective is to provide cleaner input data for training generative models.
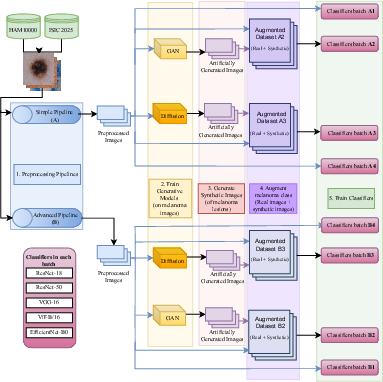
Figure 2: Overall Design of the experimental study.

Figure 3: Stages of the Dullrazor algorithm. Stages of the algorithm are: (a) Original image with artifacts. (b) Blackhat mask. (c) Binary mask. (d) Final image free of artifacts.
StyleGAN2-ADA uses a style-based architecture to generate high-fidelity images through adversarial training, while DDPMs are deployed to generate diverse samples by iteratively denoising a latent space representation. Each generative model was trained on datasets processed through both pipelines, and the synthetic images were used to augment existing melanoma datasets. These augmented datasets were then employed to train deep learning classifiers, including ResNet18, ResNet50, VGG16, ViT-B/16, and EfficientNet-B0, to evaluate the impact of synthetic data on melanoma diagnosis.
Results
The results demonstrated that the choice of generative architecture significantly impacts image fidelity and diagnostic utility, with StyleGAN2-ADA achieving superior results in terms of Fréchet Inception Distance (FID) and Kernel Inception Distance (KID), indicating a closer alignment with real data distributions. Conversely, DDPMs generated images with higher variance and diversity but at the cost of perceptual fidelity and class anchoring.
Evaluation of preprocessed and synthetic data via t-SNE embeddings further revealed distinct clustering patterns (Figure 4), confirming the superiority of GAN-generated images in maintaining class coherence. Advanced preprocessing only yielded marginal improvements, suggesting that aggressive artifact removal could suppress clinically relevant texture cues.

Figure 4: t-SNE embeddings for the basic (left) and advanced (right) pipelines, showing the distributions of GT, GN, and DF samples.
The incorporation of synthetic data resulted in significant improvements in melanoma detection, with F1-scores increasing by 8–15% and ViT-B/16 achieving an F1-score of approximately 0.88 and ROC-AUC of approximately 0.98, representing a 14% improvement over baseline models without synthetic augmentation.
Discussion
The findings highlight that augmenting dermoscopic datasets with GAN-generated synthetic images substantially benefits classification models, particularly for underrepresented classes like melanoma. Transformer models, specifically ViT-B/16, exhibited robust performance improvements, cementing their role in contemporary diagnostic pipelines.
Further analysis of visual interpretability through Grad-CAM highlighted inherent differences in classifier focus, with ViT-B/16 exhibiting diffuse activation patterns potentially attributable to attention-based image interpretation. These results underscore the potential of GAN-based augmentation strategies in clinical dermatology, providing a balanced improvement across realism, diversity, and diagnostic performance.
Conclusion
The study effectively establishes SkinGenBench as a robust benchmark for assessing generative models and preprocessing methodologies in synthetic dermoscopic image augmentation. With implications extending to clinical melanoma detection, the research affirms the need for careful selection of preprocessing and generative strategies to maximize image fidelity and diagnostic accuracy. Future work may explore more advanced diffusion models and incorporate multi-institutional datasets to further generalize findings across diverse populations.





