- The paper introduces a unified multi-task generative model using flow matching to enhance de novo ligand design, docking, and interactive pharmacophore-guided processes.
- It extends geometric graph neural networks to support heterogeneous molecular graphs and complex conditioning, achieving up to 91% docking accuracy and significant interaction recovery improvements.
- Empirical evaluations show state-of-the-art physical plausibility in design tasks while highlighting that transfer learning benefits across tasks remain modest.
OMTRA: Multi-Task Generative Modeling for Structure-Based Drug Design
Introduction and Motivation
OMTRA introduces a unified framework for structure-based drug design (SBDD), leveraging multi-modal flow matching to accommodate multiple generative and predictive tasks relevant to protein–ligand modeling. The paradigm shift from task-specific approaches to a single multi-task architecture enables both de novo ligand generation and docking, as well as tasks with no analogue in classical SBDD such as interactive pharmacophore-guided design. OMTRA's flexibility is attributed to its capacity to arbitrate between generated and conditioning modalities, supporting ligand, protein pocket, pharmacophore, cofactors, ions, and extensions to post-translational modifications.

Figure 1: OMTRA overview—multi-task generative flow matching of ligands conditioned on protein pockets and pharmacophores, supporting de novo design, docking, and conformer generation.
The model is underpinned by a curated dataset of 500M 3D molecular conformers, augmenting the protein–ligand and pharmacophore data landscapes. This scale not only expands accessible chemical diversity but also facilitates pretraining in protein-free domains, addressing bottlenecks common in transfer learning across molecular modalities.
OMTRA extends the FlowMol3 geometric graph neural network architecture to operate over heterogeneous graphs comprising ligand, protein, and pharmacophore nodes and edges. Each node embeds continuous spatial coordinates, scalar features (condensed atom types), and vector features; edge attributes are tailored for ligand–ligand and other–other interactions. Modality-specific SE(3)-equivariant convolutions and GVP blocks drive message passing, with parameter sharing across tasks enforced when operating in multi-task configuration.
The generative process is encoded in multi-modal flow matching: continuous modalities (3D positions) are evolved under ODEs parameterized by neural nets trained to denoise intermediate samples; discrete modalities (atom types, bond orders, pharmacophore types) are treated with masked discrete flow matching via continuous-time Markov chains. The joint loss aggregates modality-specific denoising and cross-entropy objectives weighted per task.
Supported Tasks and Data Curation
OMTRA supports arbitrary partitions over modalities, allowing for:
- Unconditional de novo ligand design
- Ligand conformer generation
- Rigid and flexible docking (optionally pharmacophore-conditioned)
- Pocket-conditioned and pharmacophore-conditioned de novo design
- Combined pocket/pharmacophore-driven ligand design
Upcoming tasks include flexible protein–ligand co-folding, joint ligand–pharmacophore generation, and fully de novo protein–ligand–pharmacophore modeling.
Datasets include the Pharmit set (500M conformers), Plinder (400K protein–ligand complexes with curated splits), Crossdocked (22.5M protein–ligand poses), and PoseBusters benchmark (428 complexes for docking validation). Molecular representations are harmonized across these sources, with condensed atom typing for ligands, SMARTS-derived pharmacophore detection, and positional as well as residue-type encoding for proteins.
OMTRA achieves state-of-the-art physical plausibility in de novo design (89.8% PB-valid) and top-1 docking accuracy (>91% PB-valid, RMSD < 2Å) surpassing prior generative models including Pocket2Mol, DrugFlow, TargetDiff, DiffSBDD, and commercial tools such as AlphaFold3, Gnina, Vina, and SurfDock. The fraction of ligands achieving native-like protein–ligand interactions is elevated by up to 37% compared to baselines.


Figure 2: PoseBusters validity comparison between OMTRA multi-task and single-task models for de novo design and docking.
Ablation studies reveal that large-scale ligand-only pretraining improves plausibility and interaction metrics for de novo design but can marginally impair docking validity, while joint multi-task training does not consistently outperform isolated single-task models for all objectives. These results suggest transfer learning effects in small molecule generative modeling are modest, challenging assumptions regarding universal parameter sharing.
Conditional Generation: Pharmacophore and Pocket Context
OMTRA exploits pharmacophore and pocket conditioning to steer structure generation.

Figure 3: Unconditional and pharmacophore-conditioned ligand trajectories highlight stepwise evolution and control over functional group placement.

Figure 4: Pocket-conditioned sampling trajectories—guiding ligand evolution directly inside binding pockets using protein and/or pharmacophore information.
When pharmacophore constraints are supplied interactively, the system achieves dramatic improvements in interaction recovery (~10 percentage points for de novo design, ~10 for docking) and pharmacophore satisfaction rates (>97%).
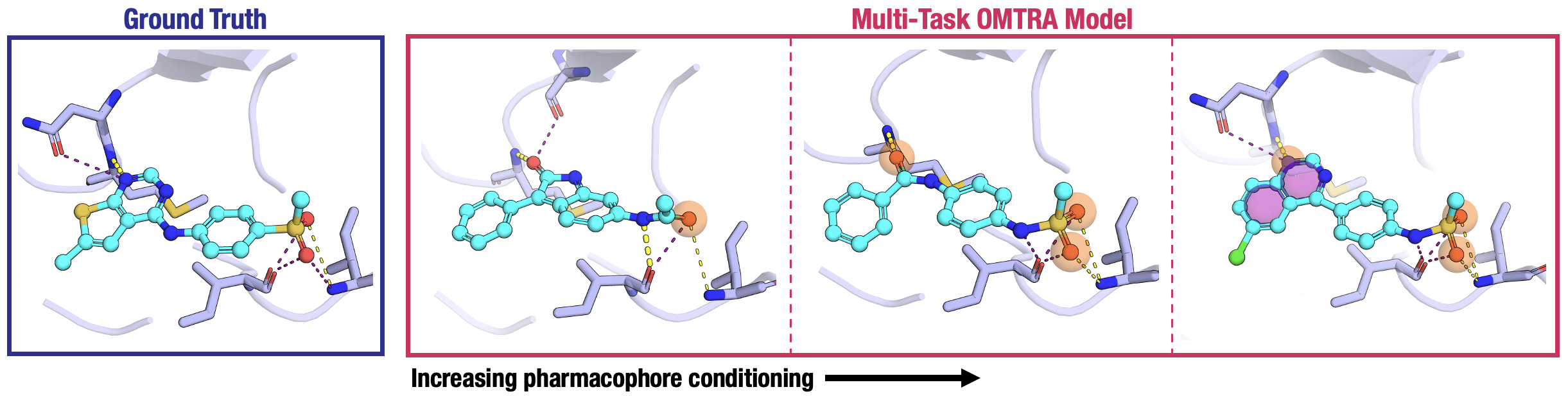
Figure 5: Pharmacophore-guided design—generated ligands matched to 1–3 pharmacophore centers anchored in the binding pocket.
The quality of both de novo design and docking increases continuously as a function of the number of supplied pharmacophore centers.

Figure 6: Bar plots showing de novo ligand quality improving with the number of pharmacophore centers used for conditioning (40,000 samples).

Figure 7: Docking success (RMSD < 2Å, interaction recovery) as a function of pharmacophore conditioning (40,000 samples).
Practical implications are clear: interactive or automated extraction of pharmacophores from known binders can be leveraged to reduce sampling requirements and enhance protein–ligand interaction matching.
Representative Outputs and Qualitative Assessment
OMTRA generates diverse ligands for targets such as thiamin phosphate synthase (PDB: 1G4S), achieving high (85.7–100%) interaction recovery for de novo samples.

Figure 8: De novo samples conditioned on thiamin phosphate synthase pocket—interaction recovery up to 100%.
PoseBusters evaluations validate geometric, chemical, and stereochemical plausibility of OMTRA outputs for both design and docking tasks.
Implications and Future Directions
OMTRA’s minimal architectural extensions enable multi-task operation in heterogeneous molecular graphs—yet transfer learning effects remain limited by the current paradigm. The inclusion of 500M conformer Pharmit pretraining and simultaneous modality generation did not yield deep synergistic improvement. The ability to condition on user-supplied (or algorithmically extracted) pharmacophore features is accentuated, particularly for reducing the computational load and targeting bespoke interaction networks.
The flexibility of modality partitioning positions OMTRA as an extensible base for further developments: direct generation of protein backbone or side-chain structure as an additional modality; full co-folding of protein–ligand–pharmacophore ensembles; differential training regimes to ameliorate learned transfer; and integration with physics-based scoring or reward guidance (see recent advances such as PhysDock [Zhang et al., 2025]).
From the standpoint of drug discovery workflow, OMTRA is immediately relevant for lead optimization, analog expansion, virtual screening, and fragment-based design—especially in settings where explicit knowledge of binding modes or interaction motifs is available or can be inferred.
Conclusion
OMTRA systematically advances multi-task generative modeling in SBDD by supporting simultaneous generation and conditioning across molecular modalities. Its unified flow matching framework achieves state-of-the-art metrics in de novo ligand design and docking, with strong interpretability via pharmacophore guidance. The empirical finding that large-scale pretraining and multi-tasking yield only modest benefits underlines the need for architectural innovation or alternative learning strategies if robust transfer is to be realized. OMTRA, together with the released Pharmit dataset, provides a scalable platform to drive future research in generalizable molecular generative modeling and interactive drug design.
Reference: "OMTRA: A Multi-Task Generative Model for Structure-Based Drug Design" (2512.05080)










