- The paper establishes a reproducible Tox21 leaderboard that restores consistent evaluation of toxicity prediction models using the original dataset.
- It employs Hugging Face Spaces for automated, API-driven assessment of various AI and machine learning models based on AUC metrics.
- The findings reveal that early models like DeepTox remain competitive, highlighting the impact of dataset modifications on benchmarking progress.
Measuring AI Progress in Drug Discovery: A Reproducible Leaderboard for the Tox21 Challenge
Introduction
The paper "Measuring AI Progress in Drug Discovery: A Reproducible Leaderboard for the Tox21 Challenge" (2511.14744) addresses the ongoing challenge in evaluating AI methodologies applied to drug discovery, specifically toxicity prediction utilizing the Tox21 dataset. The paper highlights the critical role played by the original Tox21 Data Challenge in 2015, which marked a significant advancement similar to the "ImageNet moment" in computer vision. The introduction of deep learning during this challenge reshaped the landscape of computational drug discovery, yet subsequent modifications to the dataset during its assimilation into various benchmarks have led to inconsistencies that obscure progress evaluation.
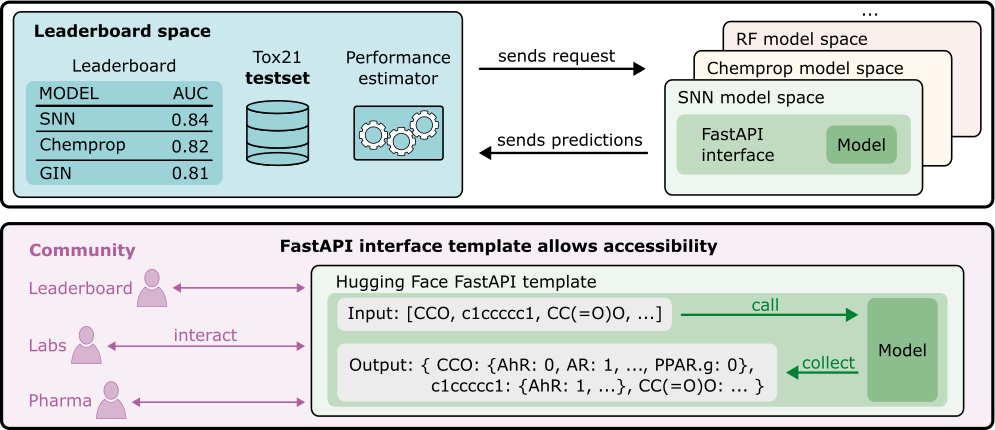
Figure 1: Overview of the Tox21 leaderboard and FastAPI interface linking model spaces with the leaderboard and external users.
Dataset Evolution and Challenges in Evaluation
The Tox21 dataset initially was a pivotal asset in toxicity prediction, encompassing endpoints related to human toxicity modeled as binary classification tasks. Its original design was altered in subsequent benchmarks such as MoleculeNet and DeepChem, where dataset splits, molecules, and labels were significantly modified. These alterations introduced comparability gaps across studies, rendering it challenging to assess progress in toxicity prediction methods. The paper proposes a solution by establishing a reproducible leaderboard hosted on Hugging Face, leveraging the original Tox21 dataset to ensure consistent evaluation of bioactivity models.
The leaderboard aligns evaluation protocols with the original dataset configuration, offering a standardized environment for submitting models that must provide predictions for all test samples. This initiative aims to restore the dataset’s integrity and facilitate a transparent, reproducible evaluation framework.
Methodology and Baseline Models
The implemented leaderboard employs Hugging Face Spaces, enabling models to be assessed through automated API-driven processes, which collate predictions from submitted models and compute standard metrics. The evaluation criteria are centered around the AUC averaged across all Tox21 targets. Notably, several traditional machine learning and AI models are re-evaluated as baselines, including DeepTox, self-normalizing neural networks (SNN), and graph neural networks (GIN).
Among the findings, descriptor-based models such as SNN show continued competitiveness, paralleling ensemble methods like DeepTox and traditional machine learning models. This persistence raises questions about the perceived progress over the decade, underscoring the necessity of re-establishing comparison frameworks for toxicity prediction.
Results and Implications
The results from the re-evaluation suggest that despite innovations in machine learning methodologies, early models like DeepTox remain competitive on the original test set. This observation indicates that while new model architectures have emerged, substantial advancement may have been hindered by the dataset and benchmark inconsistencies. Evaluations from the leaderboard differ slightly due to computational environments and methodologies, yet demonstrate consistent findings regarding model performance.
This reinstatement of faithful benchmarking can serve as a template for other molecular datasets experiencing benchmark drift. By offering a reliable structure for toxicity models, the paper contributes to enhancing AI's role in drug discovery while advocating for further investigations into few-shot and zero-shot learning evaluations.
Future Outlook
The infrastructure and findings inform future discourse on AI model evaluations in drug discovery, suggesting continued exploration into advanced learning paradigms and foundational models for molecular prediction tasks. As the landscape evolves, the paper provides guidance for systematic assessments, promoting sustained strides in AI-driven drug discovery through robust benchmarks and data integrity.
Conclusion
The paper successfully restores evaluation consistency for the Tox21 dataset, presenting a standardized framework for assessing toxicity prediction methods. This effort elucidates the trajectory of AI advancements in drug discovery, advocating for the re-evaluation of existing assumptions about method effectiveness. Through its automated, open benchmarking pipeline, the work facilitates meaningful progress review while stimulating discourse on extending current methodologies to future bioactivity predictions.
