- The paper introduces CM², a copula-driven multimodal framework that models joint distributions to capture higher-order interactions across diverse modalities.
- It employs Gaussian mixture modeling for each modality’s marginal distribution and uses variational inference to robustly impute missing data.
- Empirical results on MIMIC datasets demonstrate up to a 3.2% improvement in AUPR over baselines, validating the effectiveness of the copula approach.
Cross-Modal Alignment via Variational Copula Modelling
Introduction and Motivation
The paper presents CM2, a copula-driven multimodal learning framework designed to address the limitations of existing fusion strategies in multimodal representation learning. Traditional approaches, such as concatenation or Kronecker product, fail to capture higher-order interactions between heterogeneous modalities (e.g., EHRs, medical images, clinical notes), resulting in suboptimal joint representations. CM2 leverages copula theory to model the joint distribution of modality-specific embeddings, enabling more expressive alignment and fusion. The framework is particularly suited for scenarios with missing modalities, as it can impute missing representations via learned marginal distributions.
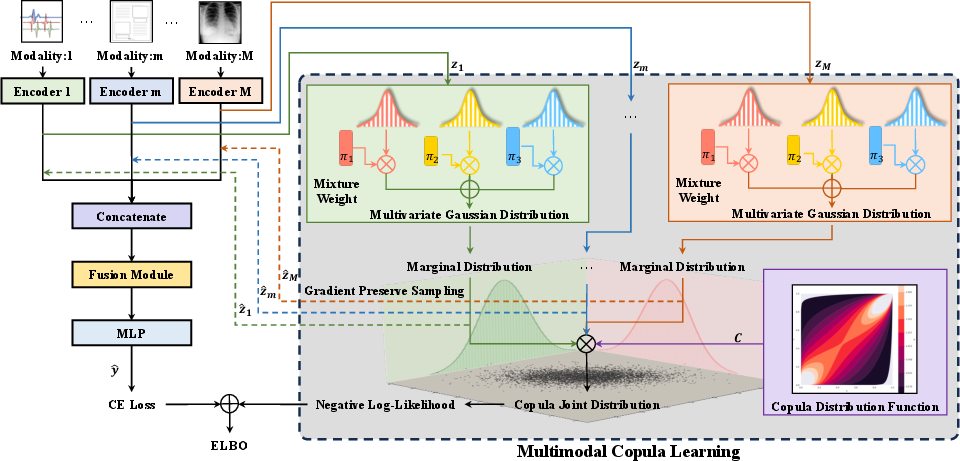
Figure 1: Overview of the CM2 framework, illustrating modality-specific embedding extraction, GMM-based marginal modeling, copula-based joint estimation, and fusion/classification pipeline.
Methodological Framework
Copula-Based Joint Distribution Modeling
CM2 interprets copula models as distribution alignment tools, grounded in Sklar's theorem, which guarantees the existence and uniqueness of a copula linking marginal and joint distributions for continuous variables. Each modality's embedding is modeled as a Gaussian mixture, providing flexibility for high-dimensional, non-Gaussian data. The joint distribution is then constructed using a multivariate copula, with the copula parameter α (or ρ for Gaussian copulas) learned via stochastic variational inference.
Marginal Modeling and Imputation
For each modality m, the marginal density is parameterized as:
fm(zm)=k=1∑KπmkN(μmk,Σmk)
where mixture weights πmk are predicted by an MLP with softmax output, and μmk,Σmk are trainable via backpropagation. Missing modalities are imputed by sampling from the learned GMM, ensuring that the generated embeddings reflect both marginal and joint dependencies.
Variational Inference and Optimization
The evidence lower bound (ELBO) objective combines the copula log-likelihood and task-specific loss (e.g., cross-entropy for classification):
ELBO=−λcopi=1∑n[logc(Q1(z1(i)),…,QM(zM(i)))−m=1∑Mlogfm(zm(i))]+Lobj
Gradients are propagated to all model parameters, including copula parameters, GMM parameters, and fusion/classification layers.
Copula Families and Interaction Modeling
The framework supports various copula families (Gumbel, Gaussian, Frank), each capturing different dependency structures. Empirical analysis demonstrates that the choice of copula affects the model's ability to capture tail dependencies and symmetry in modality interactions.
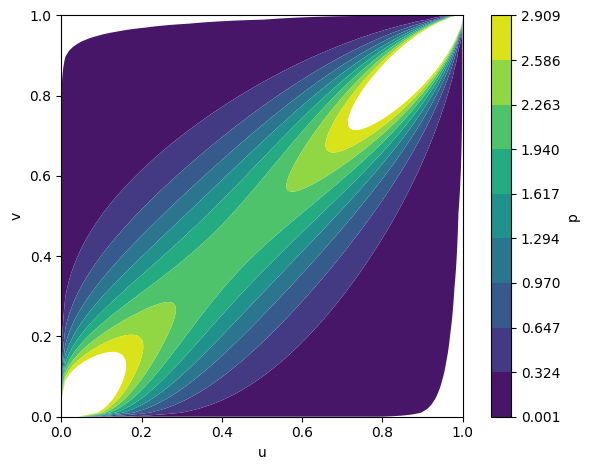
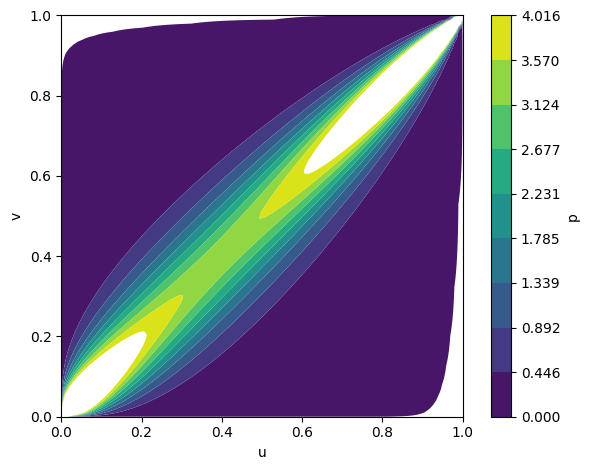
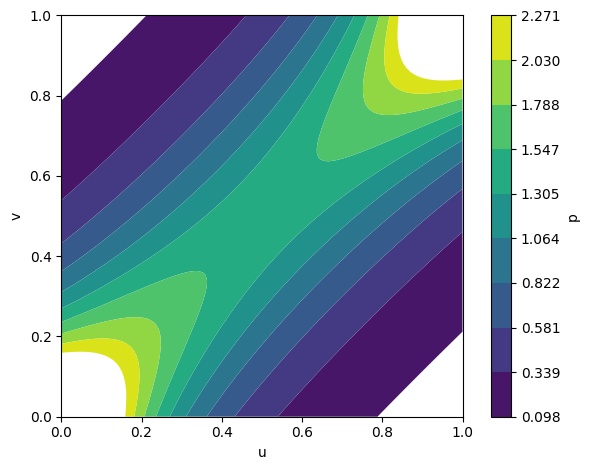
Figure 2: Fitted copula densities for Gumbel, Gaussian, and Frank families, illustrating distinct inter-modality dependency structures.
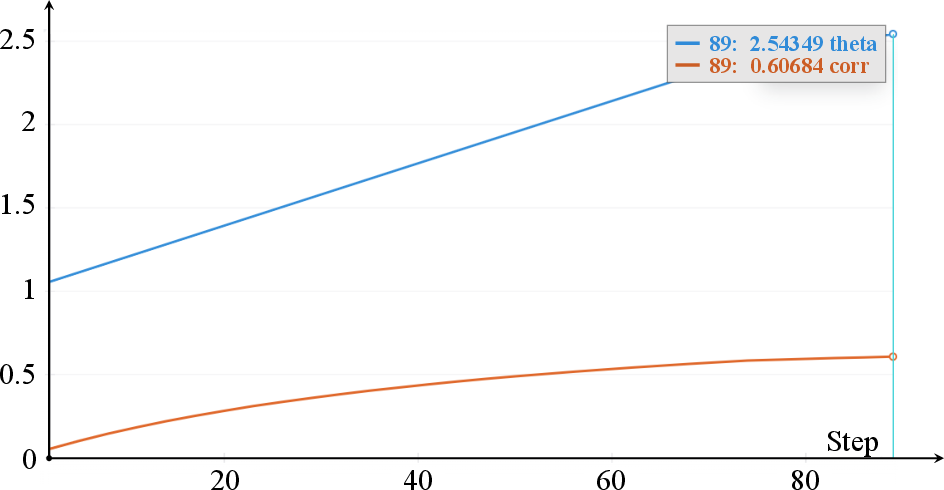
Figure 3: Evolution of the Gumbel copula parameter α and the induced correlation Corr=αα−1 over training epochs.
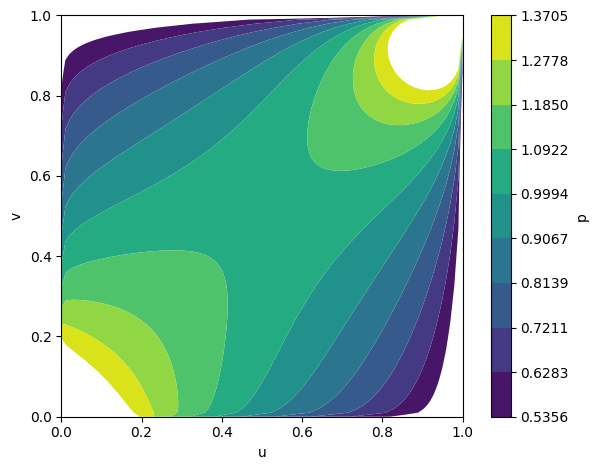
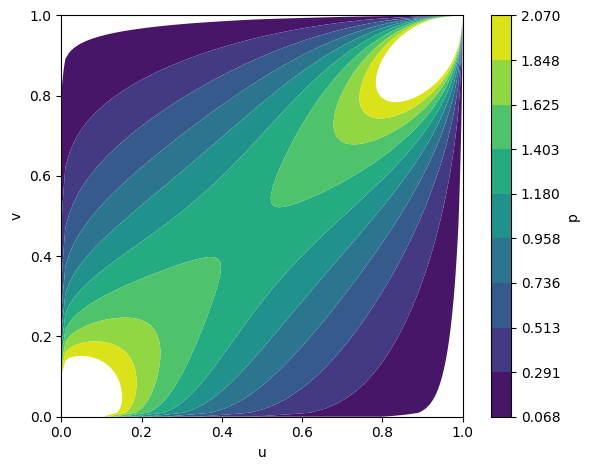
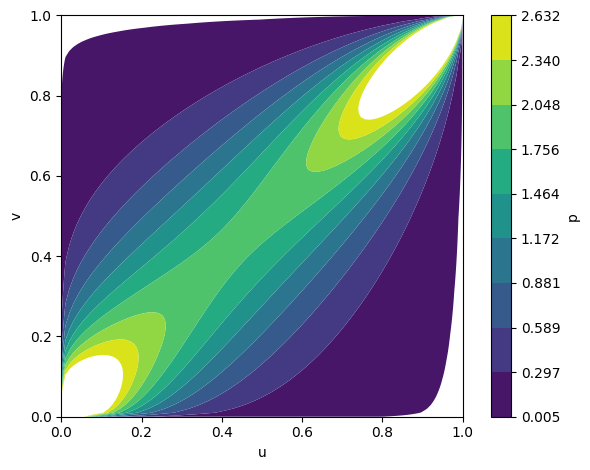
Figure 4: Temporal evolution of Gumbel copula densities at epochs 5, 50, and 100, showing progressive learning of positive dependence.
Empirical Evaluation
Datasets and Experimental Setup
Experiments are conducted on MIMIC-III, MIMIC-IV, and MIMIC-CXR datasets, encompassing EHR time series, chest X-ray images, clinical notes, and radiology reports. Both fully matched and partially matched (missing modality) scenarios are evaluated. Backbone encoders include ResNet34 for images, LSTM for time series, and TinyBERT for text.
Quantitative Results
CM2 consistently outperforms competitive baselines (MMTM, DAFT, Unified, MedFuse, DrFuse) in AUROC and AUPR across both IHM and READM tasks. Notably, CM2 achieves up to 3.2% improvement in AUPR over the best baseline on MIMIC-IV. The framework demonstrates robustness to missing modalities, with superior performance in partially matched settings due to effective imputation and joint modeling.
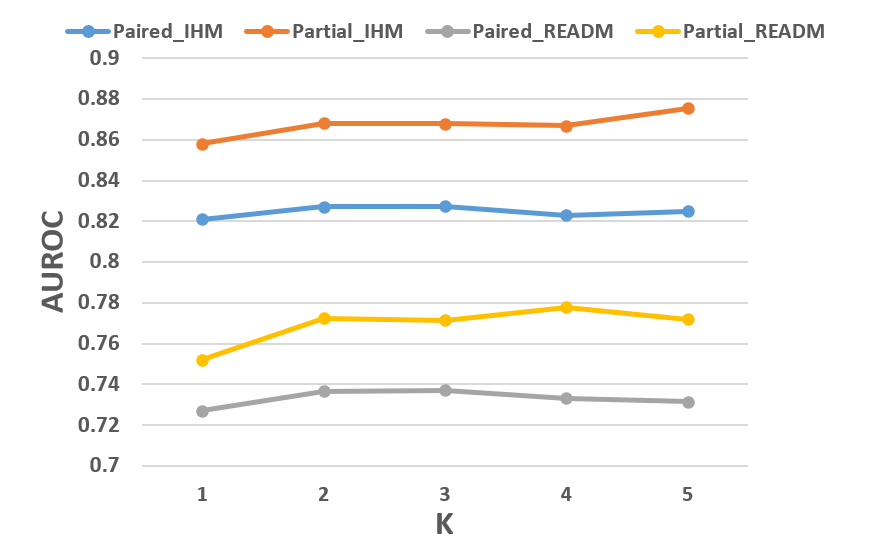
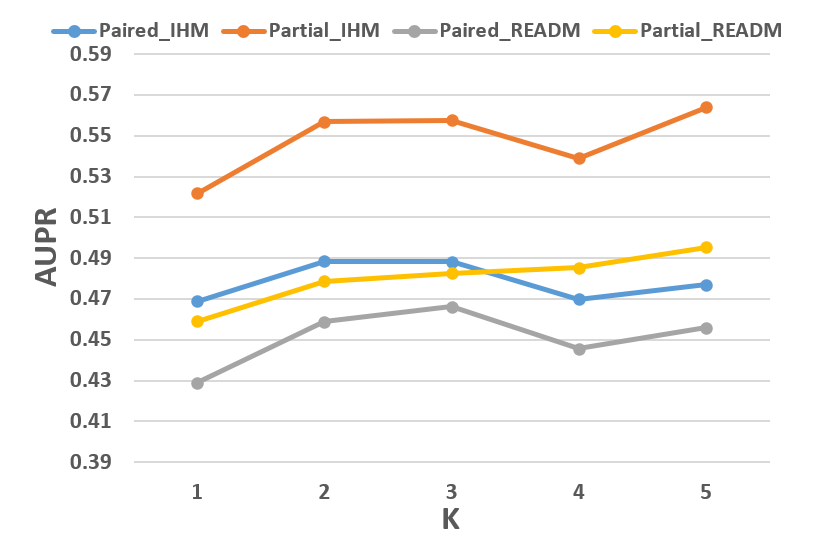
Figure 5: AUROC and AUPR results of CM2 on MIMIC-IV, highlighting performance across different numbers of GMM mixtures.
Ablation and Sensitivity Analyses
- Alignment Loss: Copula-based alignment outperforms cosine similarity and KL divergence.
- Module Contribution: Removal of copula alignment or fusion module leads to significant performance degradation, underscoring their necessity.
- Copula Family Choice: Performance is robust across copula families, but optimal choice is task-dependent.
- Scalability: Stochastic variational inference enables training on large-scale datasets with moderate computational overhead (single RTX-3090 GPU, batch size 16–32).
Theoretical and Practical Implications
The use of copula modeling in multimodal learning provides a principled approach to joint distribution estimation, with theoretical guarantees from Sklar's theorem. The framework is extensible to additional modalities and can be adapted for other tasks requiring distribution alignment, such as domain adaptation and multi-view learning. The probabilistic imputation mechanism is particularly valuable in clinical and real-world settings with incomplete data.
Limitations and Future Directions
The non-convexity of the joint log-likelihood with respect to copula parameters may hinder optimization; future work could explore alternative algorithms (e.g., partial likelihood) for improved convergence. Extension to other domains and modalities, as well as integration with transformer-based fusion architectures, are promising avenues.
Conclusion
CM2 introduces copula-based joint modeling to multimodal representation learning, enabling expressive alignment and robust handling of missing modalities. Empirical results validate its superiority over existing methods, and ablation studies confirm the critical role of copula alignment. The framework is theoretically grounded, practically scalable, and extensible to broader multimodal and distribution alignment tasks in machine learning.









