- The paper introduces FuseCPath, a framework that fuses heterogeneous foundation models to enhance whole slide image analysis.
- It employs multi-view spectral clustering, patch re-embedding transformers, and attention-based aggregation for efficient feature integration.
- Experimental results on TCGA datasets show improved biomarker prediction and survival analysis performance over traditional methods.
Fusion of Heterogeneous Pathology Foundation Models for Whole Slide Image Analysis
The paper "Fusion of Heterogeneous Pathology Foundation Models for Whole Slide Image Analysis" proposes a novel framework, FuseCPath, designed to enhance whole slide image (WSI) analysis by leveraging heterogeneous pathology foundation models (FMs). The key idea is integrating both patch-level and slide-level FMs to improve prediction accuracy and interpretability in computational pathology tasks. This essay provides a detailed examination of the framework, its components, and its empirical evaluations.
Overview of FuseCPath Framework
The FuseCPath framework is innovative in its approach to combining heterogeneous foundation models to enhance WSI analysis. Traditional approaches like separately utilizing patch-level or slide-level FMs often neglect the complementary information that could be obtained from simultaneous fusion. The FuseCPath framework uses ensemble learning concepts to fuse information at both levels, aiming to enhance performance across multiple pathology-related tasks.
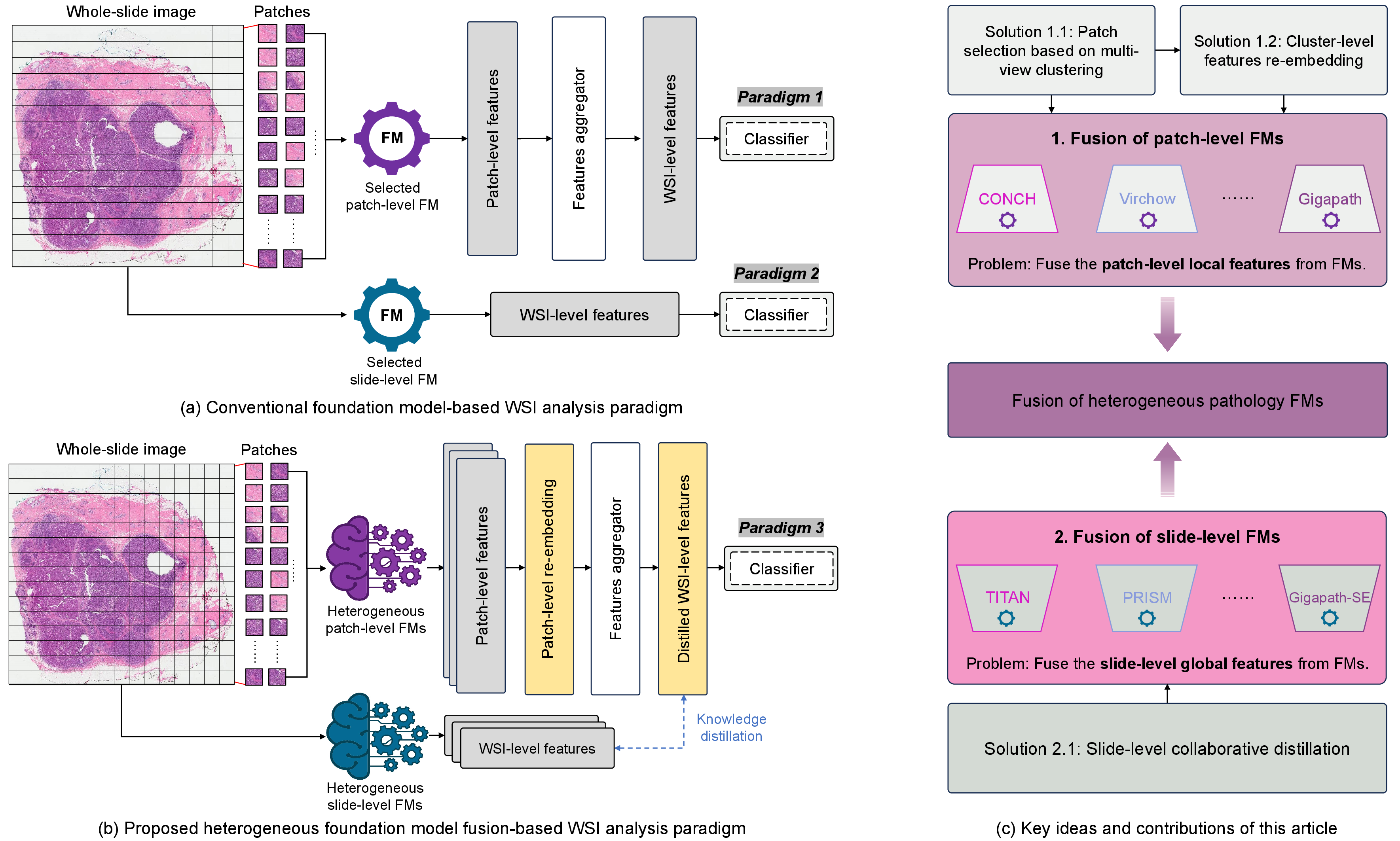
Figure 1: The proposed fusion framework compared to traditional models, showcasing the simultaneous utilization of heterogeneous patch-level and slide-level FMs.
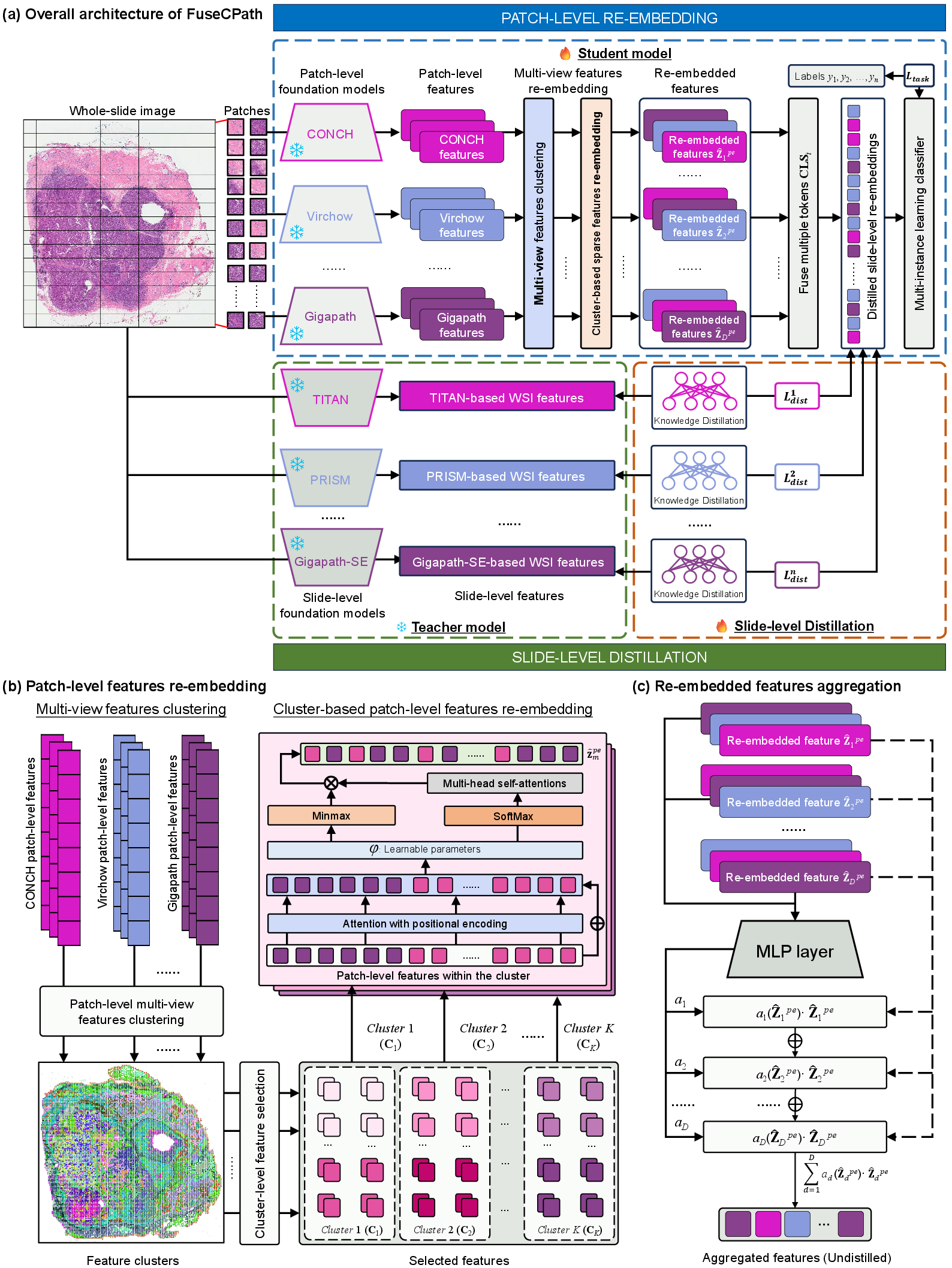
Figure 2: Detailed architecture of the FuseCPath framework, illustrating the main components such as patch-level features re-embedding and slide-level features collaborative distillation.
Multi-View Patch Features Clustering
The framework employs a multi-view spectral clustering method to select representative patches from WSIs. This method overcomes the limitations of random selection by ensuring that the chosen patches contribute significantly to the training and are representative of the WSI dataset.
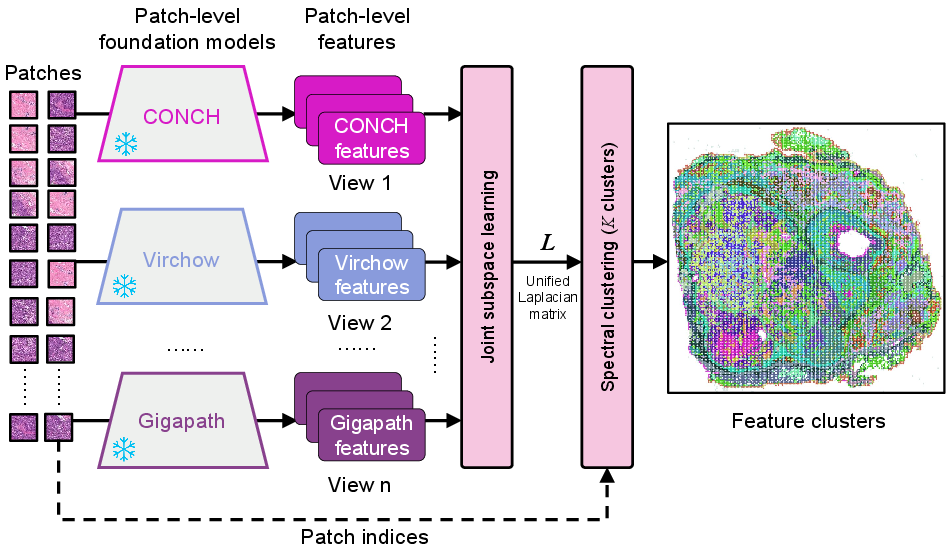
Figure 3: Multi-view spectral clustering process for integrating diverse patch embeddings as views to derive representative patches.
This clustering reduces computational load while preserving critical information, a crucial step given the high resolution of WSIs, which makes processing in GPU confined environments challenging.
Patch-Level Features Re-Embedding and Aggregation
Patch-level feature re-embedding is handled through a novel Cluster-level Re-embedding Transformer (CR2T), which fuses patch embeddings from different models into a cohesive set of features. The re-embedding procedure focuses on online capturing of local features through advanced attention mechanisms like Regional Multi-head Self-attention, ensuring efficient computational performance.
The aggregation of these re-embedded features uses Attention-Based Multiple instance learning (AB-MIL), which combines the re-embedded patch features into a unified feature set for further analysis.
Slide-Level Collaborative Distillation
FuseCPath introduces a collaborative distillation strategy to bridge the dimensional gap between patch- and slide-level features. This is achieved by considering slide-level features from gigapath-based models as teacher outputs, which guide the training of student models representing patch-level characteristics. Such a strategy ensures that global features assist in refining local feature representations, thus enhancing the model's robustness and accuracy across tasks.
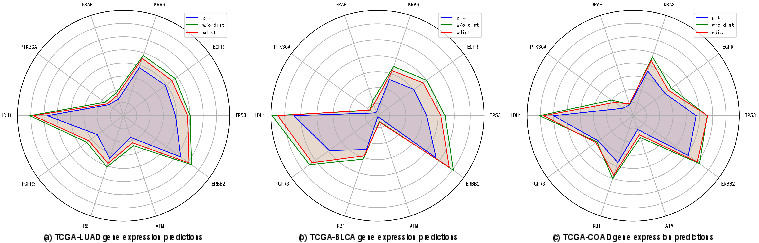
Figure 4: Gene expression prediction visualizations, highlighting the approach's closeness to ground truth values.
Experimental Results and Implications
Extensive experiments conducted on TCGA datasets illustrate that FuseCPath achieves superior performance in tasks such as biomarker prediction, gene expression analysis, and survival analysis. For instance, FuseCPath demonstrated a marked improvement in AUROC metrics across several biomarker prediction tasks compared to traditional methods and single-model approaches.
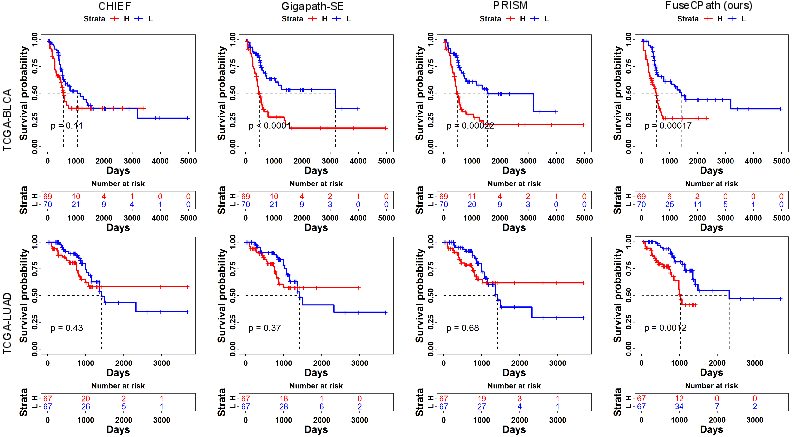
Figure 5: Kaplan-Meier survival curves illustrating improved stratification performance when using FuseCPath.
The framework's capacity to integrate diverse information from heterogeneous FMs is shown to contribute significantly to prediction accuracy and stability.
Conclusions
The paper presents a compelling case for integrating multiple foundation models in WSI analysis to leverage their complementary strengths. FuseCPath's unified approach to handling both patch-level details and slide-level overview enhances predictive performance, offering a powerful tool in computational pathology. Its innovative use of multi-view clustering, re-embedding transformers, and collaborative distillation provides a robust pathway forward in integrating diverse FM capabilities. This work opens avenues for extending model fusion strategies to include additional modalities, potentially enhancing the scope and accuracy of digital pathology analyses.




