- The paper introduces CRISP, a method that persistently unlearns unwanted knowledge from language models using sparse autoencoders.
- It utilizes a two-phase process with contrastive activation analysis for feature selection and LoRA-based fine-tuning to suppress target features while preserving benign ones.
- Experimental results on WMDP benchmarks demonstrate that CRISP outperforms existing unlearning methods with significant gains in unlearning and retention metrics.
Persistent Concept Unlearning via Sparse Autoencoders
Introduction
The paper introduces CRISP (Concept Removal via Interpretable Sparse Projections), a parameter-efficient methodology designed to persistently unlearn specific knowledge from LLMs using Sparse Autoencoders (SAEs). As LLMs continue to be implemented in real-world applications, the demand to erase certain knowledge—due to safety, privacy, or copyright concerns—grows. Unlike previous methods that often operate at inference time, CRISP ensures changes in the model's parameters, making them less susceptible to reversal by adversarial entities.
Methodology
CRISP operates in a two-phase process:
- Feature Selection:
- Contrastive Activation Analysis: CRISP leverages pre-trained SAEs to identify features frequently activated by the target (unwanted) corpus but not the benign (retained) corpus (Figure 1). For every feature, two primary metrics are computed:
- Activation Count Difference: Quantifies the difference in activation frequency between target and benign datasets.
- Relative Activation Ratio: Measures the feature's relative prominence in the target dataset.
- Salient Features Extraction: By applying thresholds on activation metrics, CRISP selects salient features for suppression.
- Model Optimization:
- Parameter-Efficient Fine-Tuning with LoRA: Incorporating a multi-faceted loss function that penalizes target feature activation while encouraging retention of benign features. It combines unlearning, retention, and coherence losses weighted to balance the three objectives.
- Layerwise Updates: Features identified in specific layers where activations are mapped and suppressed efficiently.
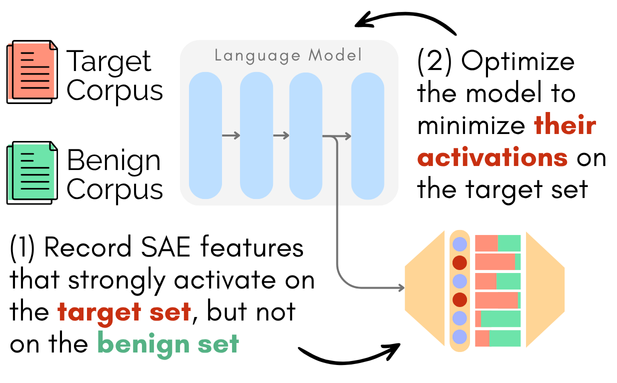
Figure 1: Overview of CRISP: (1) We identify features that are frequently and strongly activated by the target corpus---but not by the benign corpus---using pre-trained sparse autoencoders (SAEs). (2) We then fine-tune the model to suppress these features on the target corpus, while preserving their activations on the benign corpus.
Experimental Setup
Datasets and Models
CRISP is evaluated on datasets from the WMDP benchmark, including domains such as biosecurity and cybersecurity, across two LLMs: Llama-3.1-8B and Gemma-2-2B. These datasets contain target and retain corpora tailored to stress-test the unlearning capabilities while preserving unrelated knowledge.
Baseline Comparisons
CRISP is compared against contemporary unlearning techniques: RMU and ELM. Unlike these methods, CRISP leverages SAE-based feature suppression, allowing for more selective, less disruptive parameter updates.
Evaluation Metrics
Performance is evaluated based on:
- Unlearn Accuracy: Lower unplanned concept retention indicates stronger unlearning.
- Retain Accuracy and MMLU: Reflects the model's capacity to preserve general and specific benign knowledge.
- Fluency and Concept Scores: Assesses the generation quality and ability to maintain relevant conceptual knowledge.
Results
CRISP demonstrates superior performance, achieving a balance of unlearning efficacy and retention across metrics, notably outperforming baselines with improvements ranging from 5 to 34 points on the WMDP benchmark. Qualitatively, CRISP maintains fluency and coherence better than alternatives which often degrade model generation quality or lead to off-topic responses.
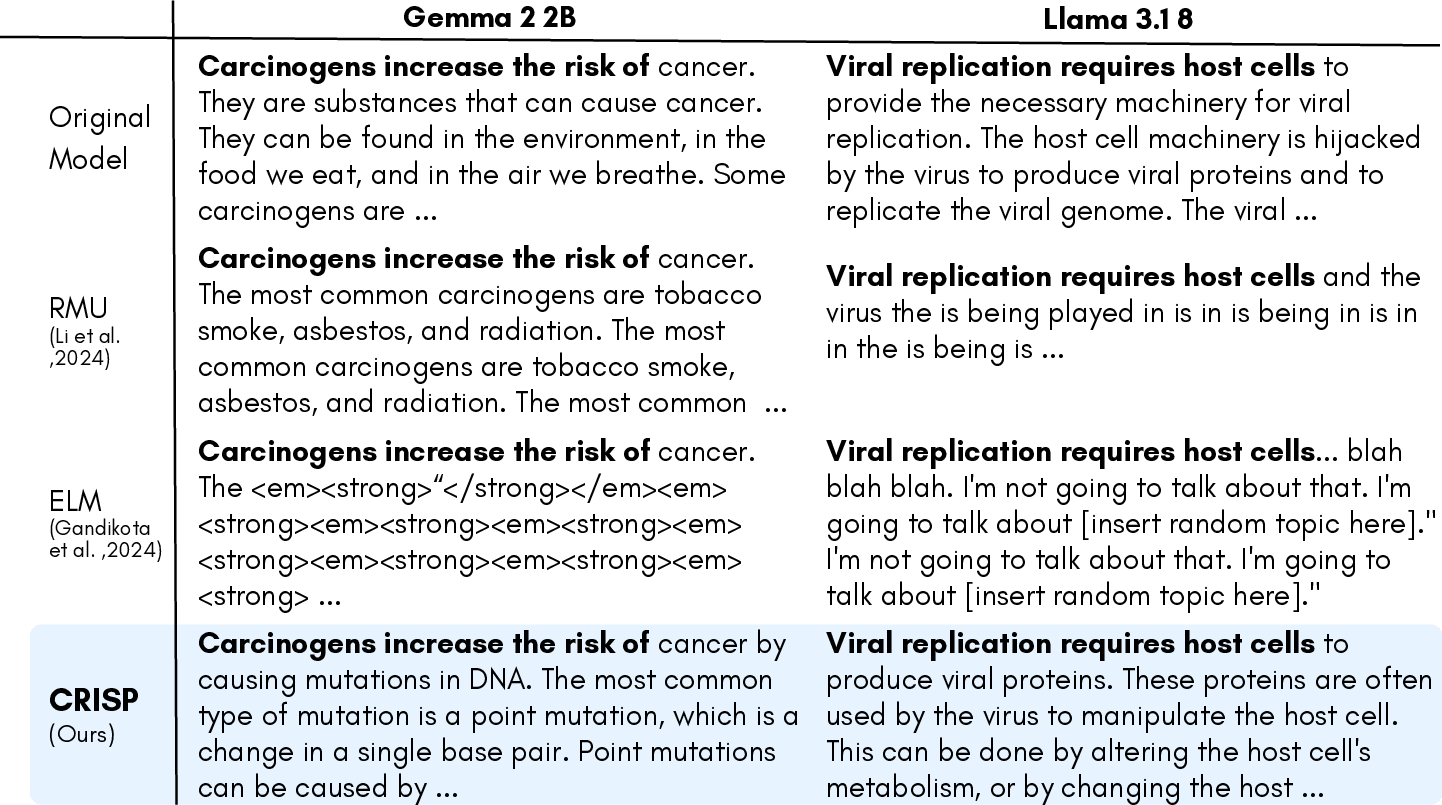
Figure 2: Qualitative comparison of generations after different unlearning methods. We prompt about non-harmful biomedical knowledge that is topically related to harmful concepts from the WMDP-Bio dataset. While existing methods disrupt fluency or inject artifacts (e.g., repetition, formatting tokens), CRISP retains coherent and informative generations, demonstrating effective preservation of general-domain capabilities.
Feature-Level Analysis
CRISP's interpretability is evidenced through feature analysis, emphasizing its precision in targeting unwanted knowledge while preserving others:
- Target Features: Predominantly characterize harmful biomedical terms, e.g., references linked to viral infections and contagion mechanisms.
- Benign Features: Encompass keywords indicative of general research and clinical terminology.
- Shared Features: Highlight non-specific linguistic tokens, demonstrating CRISP's capability to avoid generalized suppression.
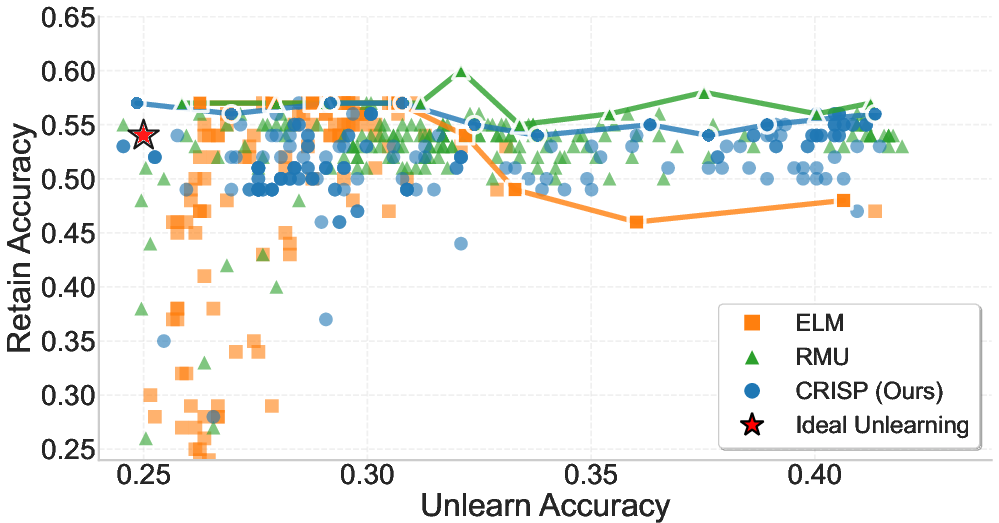
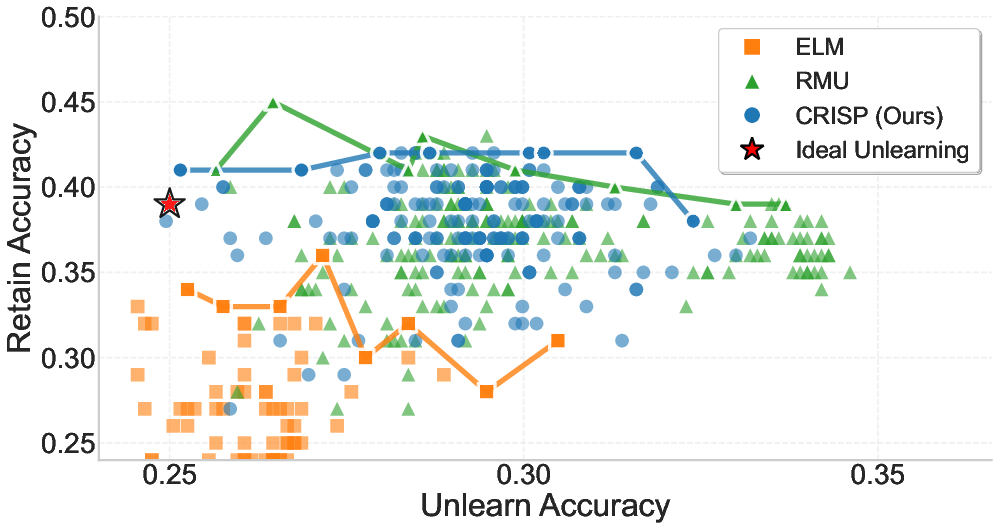
Figure 3: Trade-off between Retain Accuracy (y-axis) and Unlearn Accuracy (x-axis) on the WMDP-Cyber benchmark. Top: Llama-3.1-8B, Bottom: Gemma-2-2B. Each point shows one of 200 hyperparameter settings per method. The red star indicates the ideal outcome—complete forgetting with no loss in retain accuracy. The solid line traces the best result per unlearning bucket, forming the Pareto frontier.
Conclusion
The introduction of CRISP sets a new standard in persistent concept unlearning in LLMs by combining interpretable SAE feature extractions with efficient parameter tuning. It achieves an optimal decommissioning of undesired knowledge while retaining fluency in general model outputs. Further research could expand its application scope, adapt it to diverse models, and evaluate its resilience against adversarial attacks in variable contexts.



