- The paper introduces CoMET, a generative medical event model showing that scaling both model and data sizes enhances zero-shot clinical predictions across 78 tasks.
- The work details a transformer-based architecture with pre-layer normalization, SwiGLU activations, and grouped-query attention trained on 115B medical events from 118M patients.
- Simulation-based inference enables flexible predictions in diagnosis, prognosis, and operational forecasting, often matching or surpassing supervised models.
Generative Medical Event Models Improve with Scale: An Expert Analysis
Introduction
This paper presents CoMET (Cosmos Medical Event Transformer), a family of decoder-only transformer models trained on an unprecedented scale of structured medical event data from Epic Cosmos. The work systematically investigates scaling laws for generative medical event models and demonstrates that scaling both model and data size yields predictable improvements in downstream clinical prediction tasks. CoMET is evaluated across 78 real-world tasks, including diagnosis, prognosis, and operational forecasting, and is shown to match or outperform task-specific supervised models in most cases, without requiring task-specific fine-tuning.
Model Architecture and Training Pipeline
CoMET leverages the Qwen2 transformer architecture, incorporating pre-layer normalization, SwiGLU activations, rotary positional embeddings, and grouped-query attention. Three model sizes were trained—CoMET-S (62M params), CoMET-M (119M), and CoMET-L (1B)—using a context window of 8,192 tokens. The training dataset comprises 115B medical events (151B tokens) from 118M patients, filtered for longitudinal completeness and clinical relevance.
Medical events are tokenized using a fixed vocabulary of 7,105 tokens, representing demographics, encounters, diagnoses (ICD-10-CM), labs (LOINC quantiles), medications (ATC), procedures (CPT), and time intervals. Tokenization strategies are adapted from ETHOS, with modifications for scale and heterogeneity.
Scaling law analysis was performed via isoFLOP grid search, fitting power-law relationships for compute-optimal model size and training tokens. The exponents (α=0.520, β=0.512) closely match those found in natural language domains, indicating proportional scaling of model size and data volume is optimal.
Inference and Simulation-Based Prediction
At inference, CoMET is prompted with a patient's tokenized history and autoregressively generates future medical events, simulating possible health trajectories. Predictions for any target (diagnosis, lab, medication, procedure, encounter type) are derived from Monte Carlo aggregation over n generated timelines, enabling flexible, zero-shot prediction for arbitrary downstream tasks.
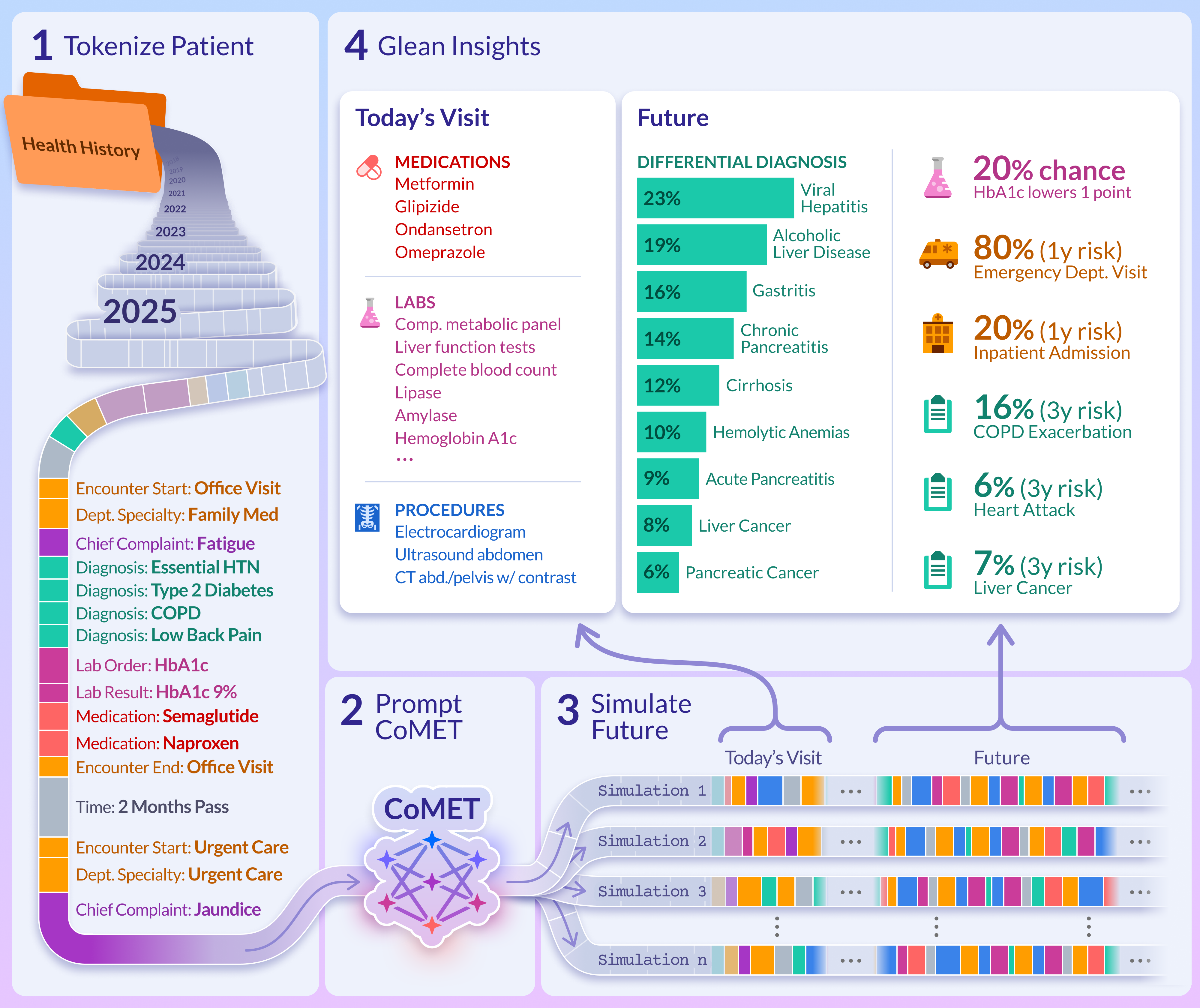
Figure 1: CoMET pretraining and inference pipeline, illustrating simulation-based prediction from patient event histories.
Evaluation: Realism and Plausibility
CoMET generations were evaluated for syntactic validity, event prevalence, and co-occurrence. Invalid multi-token event rates were <0.01% for CoMET-L, and prevalence/co-occurrence RMSLE values decreased with scale, indicating improved realism. Calibration plots for encounter frequency show good agreement between predicted and observed distributions, with expected calibration error (ECE) improving as model size increases.
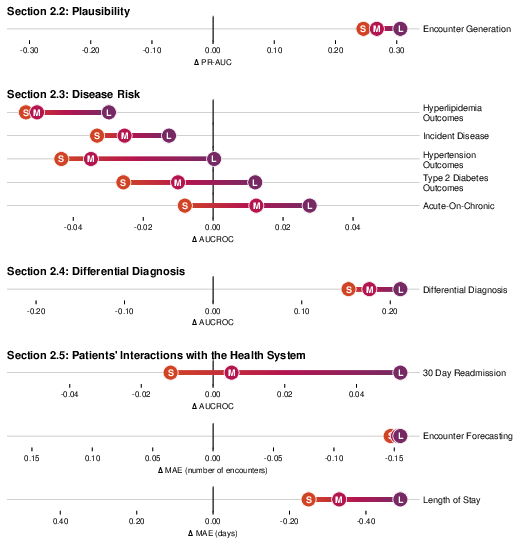
Figure 2: CoMET evaluation performance across major categories, showing improvement with scale and competitive results versus task-specific supervised models.
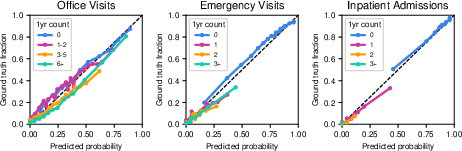
Figure 3: Calibration of CoMET-L for predicting encounter frequency by type, demonstrating accurate probabilistic forecasting.
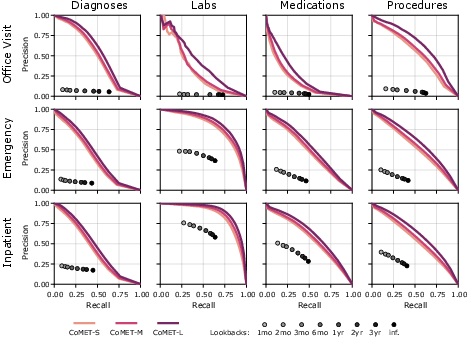
Figure 4: Precision-recall curves for single-encounter event generation, with CoMET outperforming lookback baselines and improving with scale.
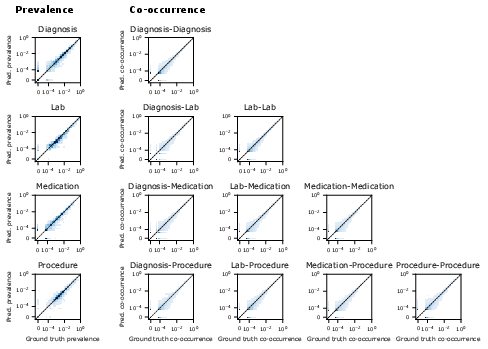
Figure 5: Heatmaps of event prevalence and co-occurrence in CoMET-L generations versus ground truth, confirming plausibility of simulated timelines.
Disease Risk Prediction and Differential Diagnosis
CoMET was evaluated on disease-specific outcomes (T2DM, HLD, HTN), acute-on-chronic events, and incident disease risk. For T2DM, CoMET-L outperformed supervised models on most outcomes (e.g., CKD progression, neuropathy, retinopathy), with AUCROC improvements up to 0.04. For hyperlipidemia, CoMET-L achieved robust AUCROC (0.93 for CHF diagnosis) but did not surpass supervised models on all tasks, indicating areas for further investigation.
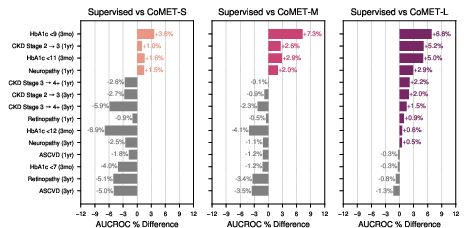
Figure 6: Percent increase in AUCROC for CoMET models versus supervised baselines on T2DM-specific outcomes.
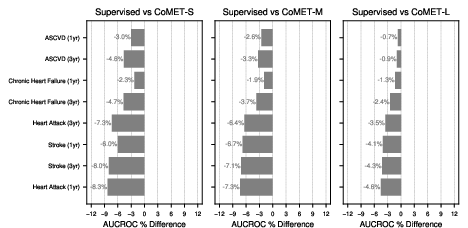
Figure 7: Hyperlipidemia-specific outcome prediction, showing consistent improvement with scale but lower performance than supervised models on diagnosis tasks.
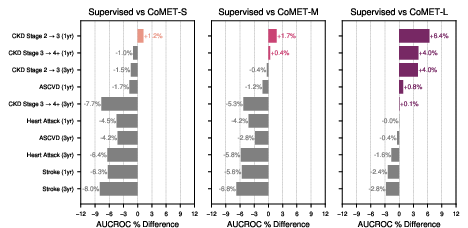
Figure 8: Hypertension-specific outcome prediction, with CoMET-L matching or exceeding supervised models on 6/10 tasks.
Acute-on-chronic event prediction (CHF, asthma, sickle cell crisis, alcohol withdrawal, COPD) showed CoMET-L outperforming baselines on 4/5 tasks. Incident disease risk prediction (COPD, CHF, dementia, asthma, alcohol use disorder, heart attack) revealed that CoMET-L achieved higher PR-AUC than baselines on all tasks, though AUCROC gains were limited by class imbalance and simulation count.
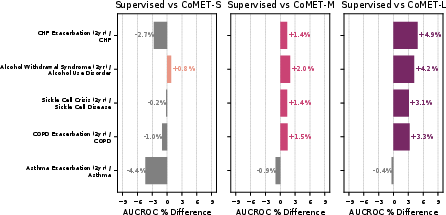
Figure 9: Acute-on-chronic outcome prediction, with CoMET-M/L outperforming baselines on most tasks.
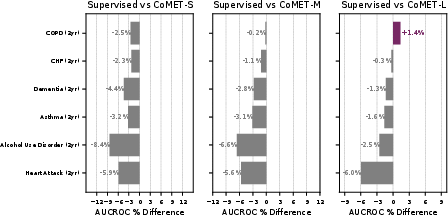
Figure 10: Incident disease risk prediction, with CoMET-L showing PR-AUC gains but limited AUCROC improvement due to low prevalence.
CoMET also demonstrated the ability to generate ranked, quantitative differential diagnoses for hepatopancreatobiliary and rheumatic disease clusters, outperforming supervised models in sensitivity and AUCROC, and flagging correct diagnoses earlier in the patient timeline.
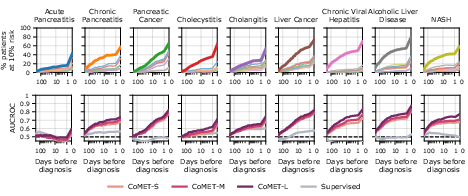
Figure 11: Hepatopancreatobiliary differential diagnosis, with CoMET-L flagging correct diagnoses earlier and with higher sensitivity.
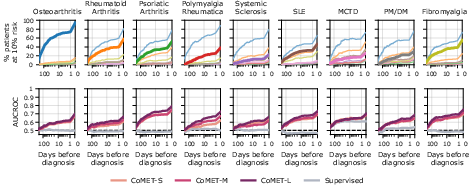
Figure 12: Rheumatic differential diagnosis, showing CoMET-L's ability to distinguish among similar presentations.
Operational Forecasting: Utilization, Length of Stay, Readmission
CoMET models were evaluated for forecasting encounter counts, hospital length of stay (LOS), and 30-day readmission risk. CoMET-L achieved lower MAE than supervised regression models for encounter frequency and LOS prediction, and higher AUCROC (0.770 vs. 0.717) for readmission risk.
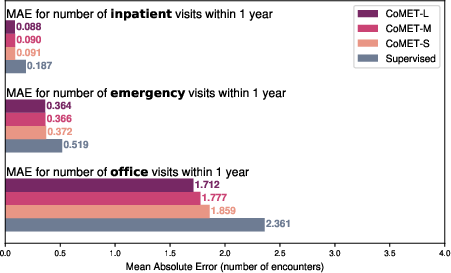
Figure 13: One-year encounter frequency forecasting, with CoMET models outperforming supervised regression baselines.
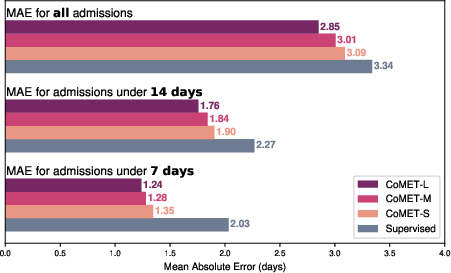
Figure 14: Hospital length of stay prediction, with CoMET models achieving lower MAE than supervised baselines.
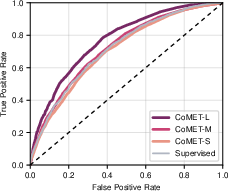
Figure 15: ROC curves for 30-day readmission prediction, with CoMET-L outperforming the best supervised model.
Scaling law analysis confirmed that training loss decreases predictably with increased model size and data volume, following power-law relationships. Downstream evaluation metrics (AUCROC, PR-AUC, MAE) improved sigmoidally as train loss decreased, with most tasks not yet plateaued, indicating substantial headroom for further scaling.
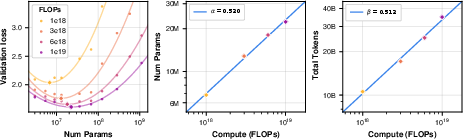
Figure 16: Scaling law analysis, showing compute-optimal model size and training tokens follow power laws.
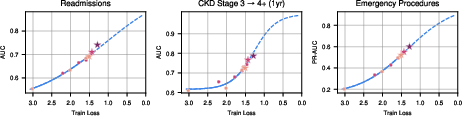
Figure 17: Downstream performance versus train loss, demonstrating sigmoidal improvement across tasks.
Test-time compute (number of generated simulations) was shown to orthogonally improve performance, especially for low-prevalence tasks, with diminishing returns beyond n≈64.
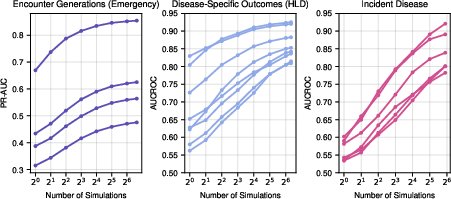
Figure 18: Effect of test-time compute on performance, with increased simulations yielding higher metrics.
Model Validity, Bias, and Fairness
Syntactic validity of generated events was high, with error rates <0.01% for CoMET-L. Subcohort analysis of AUCROC by demographic group showed no evidence of systematic bias, though further work is needed for prospective validation and fairness assessment.
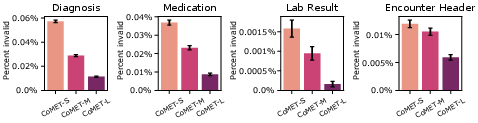
Figure 19: Syntactic validity of generated events across CoMET models.
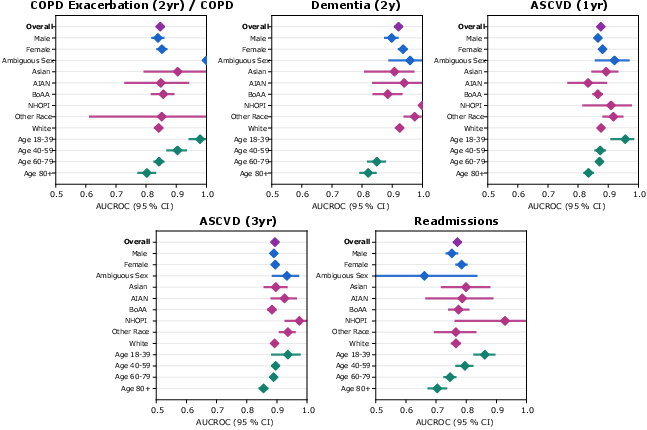
Figure 20: Subcohort analysis of bias and fairness, with CoMET-L showing consistent AUCROC across demographic groups.
Limitations and Future Directions
Key limitations include reliance on real-world EHR data subject to documentation errors and missingness, discretization of continuous values in tokenization, and context window constraints. Evaluation focused on aggregate metrics; subpopulation calibration and prospective validation remain open areas.
Future work should incorporate additional structured and multimodal data types, enable counterfactual reasoning, extend time-to-event analysis, and explore fine-tuning for task-specific improvements. Human factors research and governance frameworks are essential for responsible deployment.
Implications and Prospects
The results establish that generative medical event models scale predictably and deliver strong zero-shot performance across diverse clinical tasks. CoMET's simulation-based approach enables flexible, out-of-the-box prediction for diagnosis, prognosis, and operational forecasting, with competitive or superior performance to task-specific supervised models. The scaling law findings provide a principled methodology for resource allocation in future model development.
Theoretical implications include confirmation that scaling laws observed in NLP domains extend to structured medical event data, with similar exponents but higher token-to-parameter ratios. Practically, CoMET offers a general-purpose engine for real-world evidence generation, supporting clinical decision-making and health system operations.
Conclusion
CoMET demonstrates that large-scale generative medical event models trained on longitudinal EHR data can match or exceed the performance of task-specific supervised models across a wide range of clinical and operational tasks. Scaling both model and data size yields predictable improvements, and simulation-based inference enables flexible, zero-shot prediction. These findings position generative medical event models as a foundational technology for scalable, personalized medicine and real-world evidence generation.



















