- The paper introduces a benchmark, CGBench, that evaluates LLM scientific reasoning in clinical genetics for evidence curation tasks.
- It leverages variant and gene curation tasks with rigorous clinical guidelines, reporting metrics such as 0.420 Precision@5 and 0.63 F1 scores.
- The study reveals limitations in nuanced evidence extraction and underscores the need for advanced prompt engineering and domain adaptation.
CGBench: Benchmarking LLM Scientific Reasoning for Clinical Genetics Research
Introduction and Motivation
CGBench introduces a comprehensive benchmark for evaluating the scientific reasoning capabilities of LLMs in the context of clinical genetics. The benchmark is constructed from expert-curated annotations in the ClinGen Evidence Repository (ERepo), focusing on two central tasks in translational genomics: variant curation and gene curation. These tasks are critical for personalized medicine, requiring precise extraction, scoring, and interpretation of experimental evidence from scientific literature. Existing benchmarks in biomedical NLP are limited by their narrow scope and lack of real-world complexity; CGBench addresses this gap by formulating tasks that closely mirror the nuanced workflows of clinical genetics research.
CGBench comprises three primary tasks:
- VCI Evidence Scoring (E-Score): Multiclass classification of evidence codes for variant-disease pairs, requiring models to assign codes based on the type and strength of evidence in a publication. The evidence code hierarchy (primary, secondary, tertiary) reflects increasing granularity and complexity.
- VCI Evidence Verification (E-Ver): Binary classification to determine whether a specific evidence code is "met" or "not met" for a given variant-disease-paper tuple.
- GCI Experimental Evidence Extraction: Structured extraction of experimental evidence relevant to gene-disease validity, including category assignment, explanation, score, and reasoning for score adjustment.
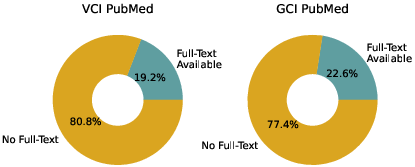
Each task is grounded in rigorous clinical guidelines (ACMG/AMP for VCI, SOPs for GCI), and the benchmark covers a diverse set of diseases, variants, genes, and evidence codes. The dataset is filtered to include only open-access full-text publications, ensuring reproducibility and transparency.
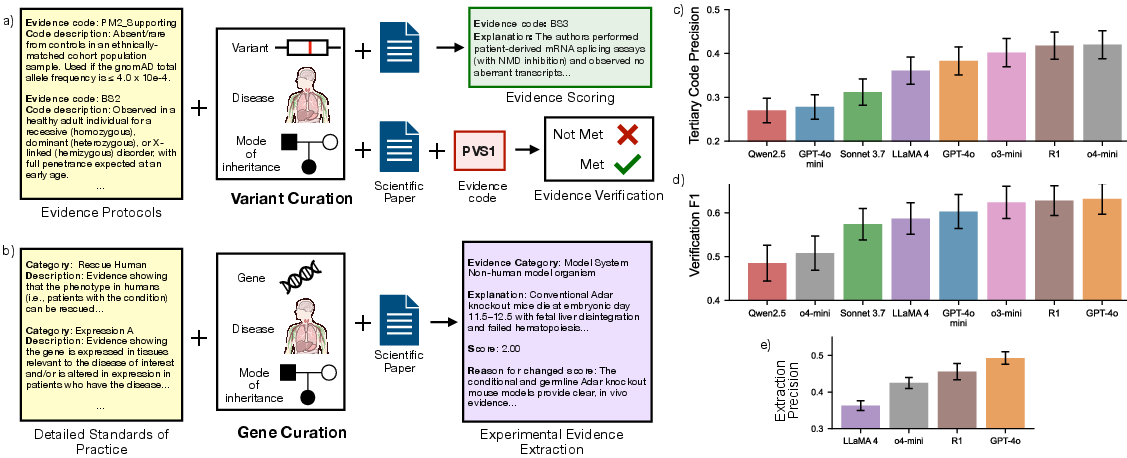
Figure 1: Overview of CGBench tasks and results, illustrating the variant and gene curation workflows and comparative model performance across evidence scoring, verification, and extraction.
Evaluation Methodology
CGBench evaluates eight LLMs, including both closed-source (GPT-4o, Claude Sonnet 3.7, o4-mini, o3-mini) and open-weight models (Qwen2.5 72B, Llama 4, Deepseek R1). Models are tested under zero-shot and in-context learning (ICL) regimes, with prompts designed to maximize task fidelity (chain-of-thought, role-playing, structured context). For evidence scoring and extraction, pass@5 sampling is used to account for stochasticity in model outputs.
Metrics include:
- Precision@5 and Recall@5 for multiclass evidence code prediction.
- True Positive Rate (TPR), True Negative Rate (TNR), and F1 for binary verification.
- Category Matching (precision/recall), Structure Adherence, Normalized MAE, and ΔStrength for evidence extraction.
- LM-as-a-judge agreement for explanation correspondence, calibrated against manual expert review.
Empirical Results
VCI Evidence Scoring
- Larger and reasoning-focused models (o4-mini, Deepseek R1) outperform smaller and non-reasoning models on tertiary code prediction, with o4-mini achieving 0.420 Precision@5.
- In-context learning improves performance, especially for fine-grained code prediction, with diminishing returns beyond 20-shot ICL.
- Model predictions for primary and secondary codes are less sensitive to paper content, suggesting reliance on prior knowledge or variant-disease associations.
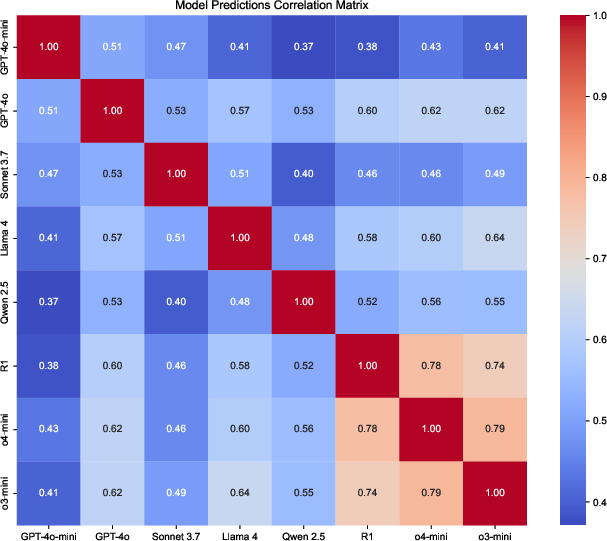
Figure 2: Prediction correlation for VCI E-Score across zero-shot models, highlighting systematic differences in model outputs.
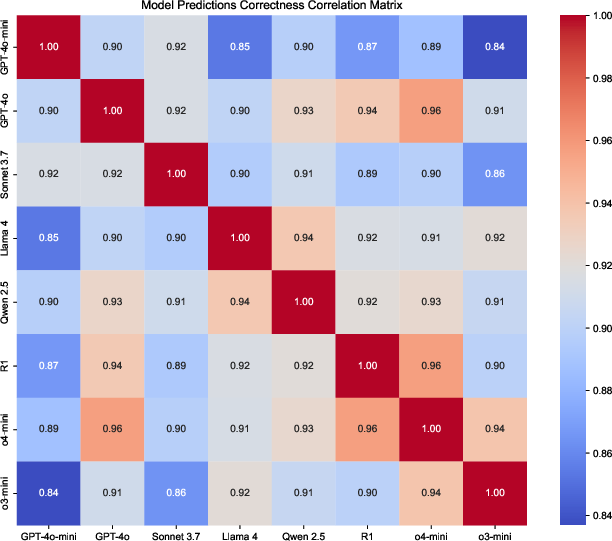
Figure 3: Correctness correlation for VCI E-Score across zero-shot models, indicating high agreement on correct predictions but divergent errors.
VCI Evidence Verification
- GPT-4o and Deepseek R1 achieve the highest F1 scores (~0.63), but overall performance is modest, with models tending to overpredict "met" status.
- In-context learning does not consistently improve verification performance, and description-only ICL is less effective than full-text demonstrations.
- o4-mini achieves the best overall extraction performance (0.186 normalized MAE, 0.445 ΔStrength), but all models over-extract evidence relative to ground-truth.
- Structure adherence is high (>95%) except for Deepseek R1, which fails to consistently produce parseable outputs.
- Score adjustment reasoning is poorly captured, with less than 50% agreement on strength changes, indicating a gap in nuanced evidence interpretation.
LM-as-a-Judge Explanation Evaluation
- Task-aware LM judge prompts yield the highest agreement with manual expert review (0.744 F1).
- In-context learning substantially improves explanation correspondence, with GPT-4o agreement rising from 0.486 (zero-shot) to 0.704 (30-shot).
- Models tend to match explanation style rather than substantive reasoning, suggesting that prompt engineering can bias outputs toward human-like explanations.
Implementation Considerations
Limitations and Future Directions
CGBench omits supplementary and multimodal data (tables, figures), which are often critical for variant interpretation. Multi-document reasoning is not addressed, and evidence prioritization remains an open challenge. The benchmark is specific to clinical genetics, and nomenclature inconsistencies may confound model evaluation. Future work should explore agentic systems for evidence synthesis, prompt-tuning for guideline adaptation, and multimodal integration for comprehensive literature interpretation.
Implications and Outlook
CGBench sets a new standard for evaluating LLMs in real-world scientific reasoning tasks, revealing substantial gaps in literature interpretation, especially for fine-grained, protocol-driven instructions. The benchmark highlights the need for advanced prompt engineering, domain adaptation, and explanation alignment to bridge the gap between automated and expert curation. As LLMs are increasingly integrated into biomedical research pipelines, rigorous benchmarks like CGBench will be essential for ensuring reliability, transparency, and clinical utility.
Conclusion
CGBench provides a robust framework for benchmarking LLM scientific reasoning in clinical genetics, exposing strengths and weaknesses across evidence extraction, scoring, and explanation. The results demonstrate that while current models show promise, significant improvements are needed for precise, guideline-driven interpretation of scientific literature. CGBench will facilitate future research in AI for translational biomedicine, driving the development of models and agents capable of expert-level evidence synthesis and interpretation.