OpenLens AI: Fully Autonomous Research Agent for Health Infomatics (2509.14778v1)
Abstract: Health informatics research is characterized by diverse data modalities, rapid knowledge expansion, and the need to integrate insights across biomedical science, data analytics, and clinical practice. These characteristics make it particularly well-suited for agent-based approaches that can automate knowledge exploration, manage complex workflows, and generate clinically meaningful outputs. Recent progress in LLM-based agents has demonstrated promising capabilities in literature synthesis, data analysis, and even end-to-end research execution. However, existing systems remain limited for health informatics because they lack mechanisms to interpret medical visualizations and often overlook domain-specific quality requirements. To address these gaps, we introduce OpenLens AI, a fully automated framework tailored to health informatics. OpenLens AI integrates specialized agents for literature review, data analysis, code generation, and manuscript preparation, enhanced by vision-language feedback for medical visualization and quality control for reproducibility. The framework automates the entire research pipeline, producing publication-ready LaTeX manuscripts with transparent and traceable workflows, thereby offering a domain-adapted solution for advancing health informatics research.
Paper Prompts
Sign up for free to create and run prompts on this paper using GPT-5.
Top Community Prompts
Explain it Like I'm 14
What this paper is about (big picture)
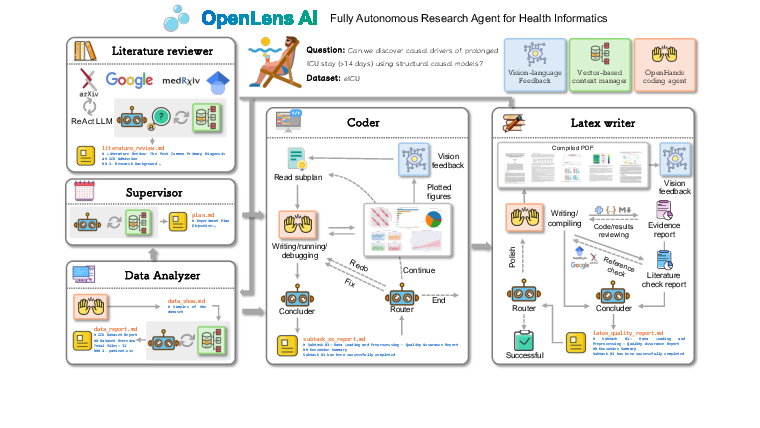
This paper introduces OpenLens AI, a computer system that can run an entire medical research project by itself. Think of it as a smart team of digital helpers that can read papers, analyze hospital data, write computer code, make charts, and even draft a scientific paper in a clean, professional format. It is designed especially for health informatics, which is the use of data and computers to improve healthcare.
What the researchers wanted to find out
In simple terms, they asked:
- Can we build a fully automatic “research agent” that handles every step of a medical paper with little or no human help?
- Can it understand medical charts and figures (not just text) and check its own work for quality and honesty?
- Will it produce research results and a full scientific paper that are clear, reliable, and ready to share?
How the system works (methods explained simply)
OpenLens AI works like a well-organized factory or a sports team where each player has a role. A user gives it a short idea (and maybe a dataset), and the system plans and completes the paper step by step. It uses several “agents” (specialized helpers):
- Supervisor: Like a project manager. It breaks the big task into smaller steps and assigns them.
- Literature Reviewer: Like a librarian-researcher. It searches online papers and writes a summary of what’s already known.
- Data Analyzer: Like a data scientist. It cleans data, runs analyses, makes charts, and explains what they mean.
- Coder: Like a programmer. It writes and fixes computer code and checks that the code actually works.
- LaTeX Writer: Like an editor. It assembles everything into a professional scientific paper, with good formatting and figures.
Two key ideas make it reliable:
- Vision-language feedback: The system doesn’t just read text; it can also “look” at charts and figures and judge if they are clear and correct. Imagine a teacher who can grade both your essay and your graphs.
- Quality control: It runs built-in checks to avoid common mistakes, like “data leakage” (using future information by accident, which is like seeing the answers to a quiz before taking it). It also links every claim in the paper back to the exact data and code used, so others can verify it.
Under the hood (in everyday terms):
- The system follows a map of steps with arrows (a workflow graph). If something fails, it retries or takes a different path, like a GPS rerouting around traffic.
- It uses tools that manage coding and keep careful notes of what happened at each step, so the whole process is traceable and repeatable.
- When it writes the final paper, it uses LaTeX (a pro tool scientists use) to produce a neat, publication-style PDF.
What they tested and what they found
They built a new set of 18 test tasks with easy, medium, and hard questions using two well-known ICU datasets (MIMIC-IV and eICU). Examples:
- Easy: What’s the age distribution of patients? What’s the mortality rate for pneumonia?
- Medium: Can we predict a patient’s 30‑day mortality from data collected in the first 24 hours? How do missing lab values affect model bias?
- Hard: What causes very long ICU stays? Do models trained in one hospital work as well on older patients or in different hospitals?
They used medium-sized AI models on purpose so hospitals could run this privately.
To grade the results, they used an “AI judge” for consistent scoring across: 1) Plan completion, 2) Code execution, 3) Result validity, 4) Paper completeness, 5) Conclusion quality.
Main results in plain words:
- For easy tasks, the system did very well. It produced correct numbers, clear charts, and tidy papers.
- For medium tasks, it was usually good but sometimes had trouble with code details or result accuracy, especially when data prep or model fitting got tricky.
- For hard tasks, it struggled more. Tough challenges like finding true causes or making models work across different hospitals and age groups led to more mistakes or weaker conclusions.
One strong point across all levels: the final papers looked professional and well-organized, with clear figures—thanks to the vision-language feedback.
Why this matters (impact and what’s next)
OpenLens AI shows that a mostly hands-off, end-to-end research assistant for healthcare is possible today. This could:
- Speed up routine analyses and first drafts of studies, saving doctors and scientists a lot of time.
- Improve trust and reproducibility by linking every claim to its data and code.
- Raise the quality of figures and the overall paper layout automatically.
However, it still has limits:
- It wasn’t directly compared head-to-head with other general systems on the same benchmark.
- Using mid-sized models may cap performance on very complex problems.
- It was tested on selected parts of two datasets; more diverse testing is needed.
The authors plan to build better public benchmarks for medical research agents, improve the models for accuracy and privacy, and make it easier to deploy in real hospitals. If successful, tools like this could help researchers focus on big ideas and patient impact, while the agent handles the heavy lifting of data, code, and writing.
Knowledge Gaps
Below is a consolidated list of concrete knowledge gaps, limitations, and open questions that remain unresolved in the paper.
- Lack of head-to-head comparisons with general-purpose research agents and human researchers on standardized, reproducible benchmarks.
- Inconsistent benchmark description (stated “18 tasks” vs. tables showing 9 tasks); unclear task accounting across datasets and duplication—needs precise, unified task specification.
- Absence of ground-truth quantitative performance reporting for predictive tasks (e.g., AUROC, AUPRC, calibration, confidence intervals); reliance on LLM-as-Judge instead of domain metrics.
- No validation of the LLM-as-Judge protocol (inter-rater reliability, agreement with expert clinicians, calibration to clinical standards); potential assessment bias unquantified.
- No ablation studies isolating module contributions (vision-language feedback, academic rigor checks, router/shared state) or sensitivity to iteration limits and model scale.
- Vision-language feedback is described qualitatively but lacks explicit evaluation criteria (e.g., checks for axes labels, units, error bars, legend consistency) and quantitative impact on error reduction.
- Use of a general-purpose VLM (GLM-4.1V-9B) without testing on medical imaging or clinical chart-specific understanding; scope appears limited to plots rather than radiology/pathology images.
- Quality control mechanisms (data leakage detection, temporal handling audits) are unspecified at the algorithmic level; coverage, false-positive/negative rates, and failure modes remain untested.
- Evidence traceability mapping from manuscript text to code/data is not formally verified (no cryptographic provenance, dataset versioning, commit hashes); reproducibility guarantees across environments are unclear.
- Code execution security, sandboxing, dependency pinning, and supply chain risk management are not discussed; risks from external packages/tools and network access remain unaddressed.
- Workflow robustness to tool/API failures, schema changes, and network outages is not evaluated (no stress/chaos testing or recovery strategies).
- Iteration caps (two refinements and two LaTeX polishes) are imposed for efficiency without analyzing their impact on final research quality or failure recovery capability.
- No reporting of runtime, compute resources, and cost per end-to-end paper; scalability to large cohorts, longer workflows, and multi-project concurrency is unquantified.
- Generalizability beyond ICU tabular/time-series data (e.g., genomics, radiology, clinical notes, multi-center EHRs) is untested; cross-specialty applicability is unknown.
- Handling of confounding and causal identification is limited; hard-task failures (SCM, generalization) are reported without methodological remedies (e.g., causal constraints, quasi-experimental designs, domain priors).
- Fairness and bias assessments (beyond age-based generalization) are absent; no subgroup analyses, equity metrics (e.g., equal opportunity, calibration by group), or bias mitigation strategies.
- Model interpretability and transparency (e.g., SHAP, partial dependence, calibration plots, decision curves) are not integrated or evaluated, especially for clinical decision relevance.
- Literature verification module is not quantitatively evaluated (metadata error rates, hallucinated citation detection/removal rates, types of citation failures).
- Manuscript quality evaluation focuses on structure/formatting; scientific rigor, novelty, and statistical correctness lack independent expert peer review.
- No human-in-the-loop safety protocols, ethical oversight, or IRB alignment; fully autonomous operation in clinical research remains risky without guardrails and review checkpoints.
- Data privacy and regulatory compliance (HIPAA/GDPR), de-identification, audit logging, and access controls for on-prem deployments are unspecified; privacy-preserving techniques are proposed but unevaluated.
- Reproducibility across runs/seeds, deterministic pipelines, environment capture (containers, package locks), and cross-system portability are not reported.
- Sensitivity to model choice, prompting strategies, router policies, and toolset variations is unknown; no cross-model evaluation (e.g., o1, R1, GPT-4o vs. GLM-4.5).
- Limited failure analysis; no taxonomy of common errors (preprocessing, modeling, evaluation), root-cause diagnosis, or targeted mitigation strategies to reduce recurrence.
- External validation and user studies with clinicians or clinical researchers are absent; user trust, adoption barriers, and workflow integration needs remain open questions.
- Use of public search tools (ArXiv, MedRxiv, Tavily) introduces risks of citation drift or retrieval errors; safeguards, audit procedures, and error correction for literature tools are not detailed.
- Benchmark scope is narrow; a larger, diverse, and standardized medical-agent benchmark with ground-truth labels and rigorous scoring rubrics (beyond emoji scales) is needed.
- No formal assessment of whether vision-language feedback improves scientific validity (not just aesthetics); measurable reductions in visualization errors or misinterpretations are not demonstrated.
- Claims of “publication-ready” LaTeX are not evaluated against journal reporting standards (e.g., CONSORT, STROBE), ethics statements, and reproducibility checklists—compliance remains unverified.
- Robustness to dataset shift and temporal drift within MIMIC/eICU is not assessed; external test sets beyond the eICU demo subset are lacking.
- Unclear minimal user input specification and safeguards to prevent spurious or clinically meaningless hypotheses; upstream hypothesis quality control is not defined.
Glossary
- Academic rigor check: An automated procedure to verify methodological soundness and prevent common scientific errors. Example: "built-in academic rigor checks to minimize hallucinations"
- Causal discovery: Methods aimed at uncovering cause-effect relationships from data rather than mere correlations. Example: "Severe issues (score1.png) appeared in causal discovery and generalization experiments"
- Comorbidity: The simultaneous presence of two or more medical conditions in a patient. Example: "What is the effect of age and comorbidity count on sepsis mortality?"
- Confound: To introduce spurious associations due to variables that influence both predictors and outcomes, biasing results. Example: "How do hospital-level differences (staff ratio, region) confound predictive modeling of mortality in eICU?"
- Data leakage: The unintended use of information during model training that would not be available at prediction time, leading to overly optimistic performance. Example: "It automatically checks for common pitfalls such as data leakage, unrealistic performance metrics, and improper handling of temporal information in time-series data."
- Directed graph-based execution structure: A workflow design where tasks are organized as nodes in a directed graph, enforcing ordered and conditional execution. Example: "enforces a directed graph-based execution structure, manages retries in case of failures, and maintains a persistent record of intermediate states."
- eICU: A multi-center critical care database used for clinical research. Example: "eICU offers a multi-center perspective covering diverse hospital settings."
- Evidence traceability check: A process linking manuscript claims to their underlying datasets, code, and logs to ensure transparency and reproducibility. Example: "Every claim or result in the manuscript is linked to its underlying evidence, including datasets, scripts, and experiment logs."
- Feature engineering pipeline: A sequence of steps to create, transform, and select input variables for modeling. Example: "It also audits feature engineering pipelines to ensure that only information available at prediction time is used."
- GLM-4.1V-9B-Thinking: A specific vision-LLM used for visual reasoning tasks in the system. Example: "GLM-4.1V-9B-Thinking \cite{hongGLM41VThinkingVersatileMultimodal2025} as the vision-LLM for visual reasoning."
- GLM-4.5-Air: A particular LLM used as the language backbone in experiments. Example: "GLM-4.5-Air \cite{teamGLM45AgenticReasoning2025} as the LLM backbone"
- ICU: Intensive Care Unit; a hospital unit providing specialized care for critically ill patients. Example: "prolonged ICU stay (>14 days)"
- LangGraph: A workflow engine for orchestrating agent steps with stateful, graph-structured control. Example: "the framework employs LangGraph\cite{LangGraph} as workflow engine that enforces a directed graph-based execution structure"
- LLM-as-Judge: An evaluation protocol where a LLM assesses the quality of system outputs along defined criteria. Example: "The outputs of the system were evaluated using an LLM-as-Judge protocol"
- MedRxiv: A preprint server for health sciences, used here as a literature search source. Example: "ArXiv, MedRxiv, Tavily"
- MIMIC-IV: A large, publicly available critical care database from a single hospital system. Example: "MIMIC-IV provides rich ICU patient records from a single hospital system"
- Missingness: The pattern and mechanism by which data entries are absent in a dataset, which can bias analyses. Example: "How do missingness patterns in lab tests influence the bias of sepsis prediction models?"
- OpenHands: A platform enabling code-centric agent execution with tool integration and state management. Example: "our coder module is built upon OpenHands\cite{wangOpenHandsOpenPlatform2024}, which provides low-level state management and seamless integration with coding tools."
- ReAct: A reasoning framework that interleaves reasoning steps with actions (tool calls) to guide retrieval and decision-making. Example: "leveraging a ReAct-based reasoning framework"
- Research-as-process automation: A paradigm where the end-to-end research workflow is decomposed and automated as a sequence of subtasks. Example: "The framework operates under the paradigm of ``research-as-process'' automation"
- Router agent: A controller that directs workflow branching by monitoring tool outputs and conditions. Example: "the router agent monitors outputs and keywords to determine whether to return for reprocessing, or proceed to final report generation."
- Sepsis: A life-threatening organ dysfunction caused by a dysregulated host response to infection. Example: "How do missingness patterns in lab tests influence the bias of sepsis prediction models?"
- Structural causal models: Graphical and equation-based models that formalize causal relationships for identification and inference. Example: "Can we discover causal drivers of prolonged ICU stay (>14 days) using structural causal models?"
- Tavily: A web search/retrieval tool used by agents for literature discovery. Example: "ArXiv, MedRxiv, Tavily"
- Traceability report: A document mapping manuscript content to supporting datasets, code, and logs to enable auditing. Example: "The system generates a structured traceability report mapping manuscript paragraphs to their supporting files and code segments."
- Vision-language feedback: The use of vision-LLMs to assess and refine visual artifacts (e.g., plots, figures) during the workflow. Example: "The system leverages vision-language feedback to ensure high-quality visual layouts and clear presentation of content."
- Vision-LLM (VLM): A multimodal model that jointly processes images and text for tasks like visual reasoning and evaluation. Example: "ensuring only VLM-approved images are included."
Collections
Sign up for free to add this paper to one or more collections.