- The paper introduces DiffSpectra, a spectrum-conditioned diffusion model that generates both 2D and 3D molecular structures from multi-modal spectra.
- DiffSpectra employs an SE(3)-equivariant Diffusion Molecule Transformer with the SpecFormer encoder, achieving a top-1 recovery of 16.01% and a top-20 accuracy of 96.86%.
- The framework overcomes traditional expert-driven limitations by ensuring robust chemical validity, with 99.9% atom stability and 98.2% molecular stability.
DiffSpectra: Spectrum-Conditioned Diffusion Models for Molecular Structure Elucidation
Introduction and Motivation
Molecular structure elucidation from spectroscopic data is a central challenge in chemistry, underpinning compound identification, synthesis, and drug discovery. Traditional approaches rely on expert-driven interpretation of spectra, which is labor-intensive and does not scale to the vast chemical space encountered in modern research. Machine learning methods have introduced retrieval-based and predictive paradigms, but these are fundamentally limited by their dependence on finite molecular libraries and their inability to generalize to novel structures. Generative models, particularly those based on SMILES or molecular graphs, have made progress but typically lack geometric inductive biases and cannot integrate multi-modal spectral information, both of which are critical for accurate structure elucidation.
DiffSpectra addresses these limitations by introducing a spectrum-conditioned diffusion framework that directly generates both 2D and 3D molecular structures from multi-modal spectra (IR, Raman, UV-Vis). The approach leverages a continuous-time diffusion process parameterized by an SE(3)-equivariant Diffusion Molecule Transformer (DMT) and conditions generation on spectral embeddings produced by a pre-trained multi-modal transformer encoder, SpecFormer.
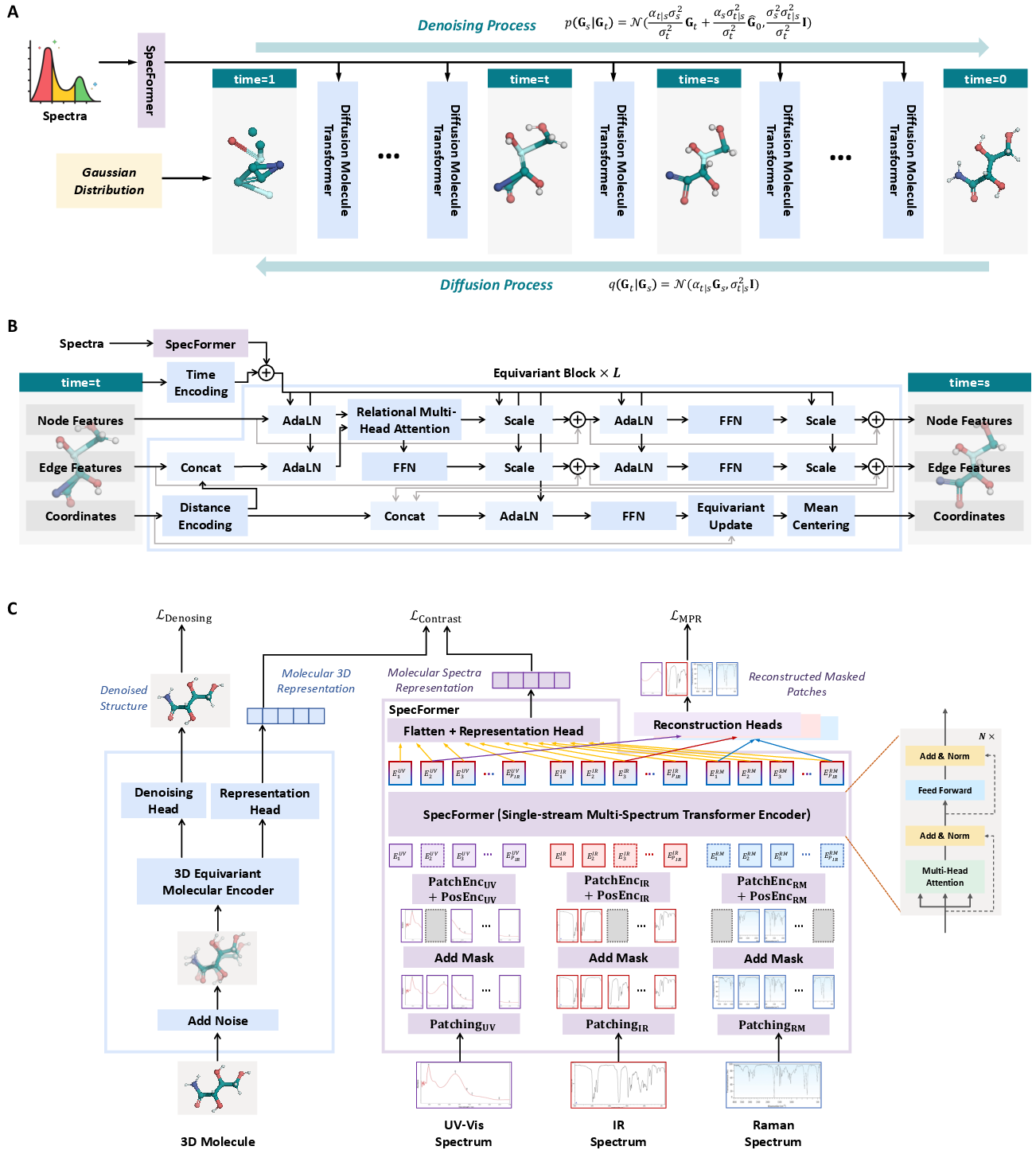
Figure 1: (A) Overview of the DiffSpectra framework, showing the forward diffusion and reverse denoising processes, the DMT denoising network, and the SpecFormer spectral encoder. (B) DMT architecture with parallel streams for node, edge, and coordinate features. (C) SpecFormer architecture and pre-training strategy for multi-modal spectra.
DiffSpectra Framework and Model Architecture
Joint 2D/3D Diffusion Modeling
DiffSpectra formulates molecular structure elucidation as a conditional generative process in a joint space of molecular graphs and 3D coordinates. Each molecule is represented as a tuple (H,A,X), where H encodes atom-level features, A encodes bond types, and X contains atomic coordinates. The forward diffusion process adds Gaussian noise to these components, and the reverse process denoises them, conditioned on spectral information.
The denoising network, DMT, is an SE(3)-equivariant transformer that processes node, edge, and coordinate streams in parallel, with extensive cross-stream interactions. This design ensures that both topological and geometric constraints are respected, and that the model is robust to rigid-body transformations.
SpecFormer is a transformer-based encoder designed to process and integrate multiple spectral modalities. Each spectrum is segmented into patches, embedded, and concatenated before being processed by a unified transformer encoder. SpecFormer is pre-trained using masked patch reconstruction (MPR) and contrastive learning to align spectral and structural representations, providing strong inductive biases for downstream conditional generation.
Training and Sampling
The model is trained using a weighted mean squared error loss over node, edge, and coordinate predictions, with SE(3)-equivariant alignment of coordinates via the Kabsch algorithm. During sampling, a temperature parameter τ modulates the stochasticity of the reverse diffusion process, allowing control over the diversity-accuracy trade-off.
Experimental Results
Molecular Generation Quality
DiffSpectra achieves high chemical validity and stability in both 2D and 3D evaluations, with atom stability at 99.9% and molecular stability at 98.2%. The model outperforms or matches state-of-the-art unconditional generative baselines (e.g., CDGS, JODO, EDM) in uniqueness, novelty, and distributional similarity metrics. In 3D geometry, DiffSpectra attains the lowest angle and dihedral MMDs, indicating accurate recovery of bond and torsional angles.
Structure Elucidation from Spectra
DiffSpectra demonstrates strong performance in spectrum-conditioned structure elucidation, achieving a top-1 exact structure recovery rate of 16.01% and a top-20 accuracy of 96.86%. Even when the exact structure is not recovered, the model produces candidates with high graph overlap (MCES), fingerprint similarity (Tanimoto and cosine), and functional group similarity (FGSim > 0.96).
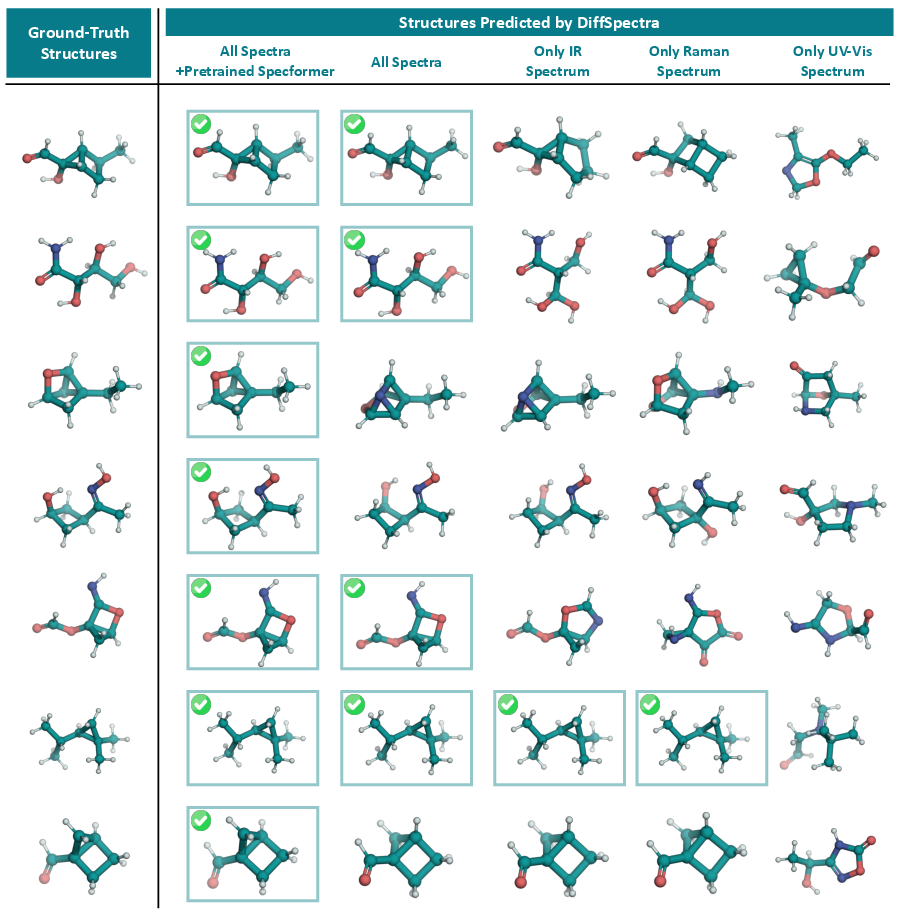
Figure 2: Visualization of structure elucidation results under different configurations: single-spectrum (IR, Raman, UV-Vis), multi-modal spectra, and with/without pre-trained SpecFormer. Ground-truth structures are shown for reference.
Ablation Studies
- Pre-training SpecFormer: Pre-training the spectral encoder yields a 2% absolute improvement in top-1 accuracy and consistent gains across all similarity metrics, confirming the value of spectral-structural alignment.
- Multi-modal vs. Single-modality Spectra: Conditioning on all three spectra outperforms any single modality, with Raman > IR > UV-Vis in isolation. UV-Vis alone is insufficient for unique structure identification in the QM9S dataset.
- Sampling Strategies: Increasing the number of samples per spectrum (top-K accuracy) dramatically improves the probability of recovering the correct structure, with top-20 accuracy approaching 97%.
- SE(3) Equivariance: Model-based SE(3)-equivariant architectures outperform data-based approaches (with or without augmentation), especially in exact structure recovery and graph overlap.
- Sampling Temperature: Moderate temperature values (τ=0.8–$1.0$) optimize the balance between diversity and accuracy; extreme values degrade performance.
Methodological Implications
DiffSpectra is the first framework to unify multi-modal spectral reasoning and joint 2D/3D generative modeling for de novo molecular structure elucidation. The integration of SE(3)-equivariant transformers and spectrum-conditioned diffusion enables the model to generate chemically valid, geometrically consistent, and spectroscopically plausible molecular structures. The use of pre-trained spectral encoders and multi-modal conditioning is shown to be critical for high-fidelity structure elucidation.
The framework is extensible to additional spectral modalities (e.g., NMR, mass spectra) and larger molecular systems, and is compatible with high-throughput experimental pipelines. The ability to generate ranked candidate lists with high recall is particularly valuable for practical applications, where downstream validation (e.g., via DFT or experimental synthesis) is required.
Theoretical and Practical Implications
The results demonstrate that spectrum-conditioned diffusion models can bridge the gap between experimental observables and molecular structure, overcoming the limitations of retrieval-based and autoregressive generative approaches. The explicit modeling of 3D geometry and the use of SE(3)-equivariant architectures are essential for capturing the physical constraints inherent in spectroscopic data.
Practically, DiffSpectra enables scalable, automated structure elucidation from routine spectroscopic measurements, with potential impact in drug discovery, materials science, and analytical chemistry. The approach is robust to the stochasticity of diffusion sampling, and the use of multi-modal spectra provides complementary structural information that enhances accuracy and reliability.
Future Directions
Key avenues for future research include:
- Scaling to larger and more diverse spectral datasets, including experimental spectra and additional modalities.
- Extending the framework to biomolecules, polymers, and crystalline materials.
- Integrating with active learning and experimental design pipelines for closed-loop molecular discovery.
- Exploring more advanced spectral encoders and generative backbones, including graph neural networks and equivariant message passing architectures.
Conclusion
DiffSpectra establishes a new paradigm for molecular structure elucidation by leveraging spectrum-conditioned diffusion models with SE(3)-equivariant transformers and multi-modal spectral encoders. The framework achieves high accuracy in recovering both 2D and 3D molecular structures from spectra, with strong empirical results and robust ablation analyses. The approach is theoretically grounded, practically effective, and extensible to a wide range of applications in chemical and materials sciences.



