- The paper introduces ATM-AMDiff, a hierarchical diffusion model that combines atom and motif views to generate drug-like molecules optimized for protein targets.
- The method leverages classifier-free guidance and equivariant graph neural networks to achieve a 98.9% validity rate in molecule generation, ensuring chemical fidelity.
- Experimental results on targets like ALK and CDK4 showcase the model's potential in de novo drug design by improving binding affinity and integrating multi-scale feature analysis.
Molecule Generation for Target Protein Binding with Hierarchical Consistency Diffusion Model
The "Molecule Generation for Target Protein Binding with Hierarchical Consistency Diffusion Model" paper introduces the ATM-AMDiff approach, a novel method for generating molecular structures designed to bind with specific protein targets. The hierarchical model combines atom-level and motif-level molecular generation to ensure high validity and novelty in identifying optimal drug candidates. This essay examines the methodology, experimental results, and implications of the AMDiff model in drug discovery.
Methodology
The AMDiff model utilizes a hierarchical diffusion approach to molecule generation, allowing simultaneous atom and motif views to construct molecular structures. The diffusion process involves two main phases: forward and reverse. In the forward diffusion, Gaussian noise is gradually added to molecule structures, while the reverse process employs a neural network to remove this noise and reconstruct viable molecular conformations.
Atom and Motif Views
The atom-view represents basic molecules using atoms as primary units, facilitating high structural diversity. It incorporates an equivariant graph neural network to predict atomic positions and types. Conversely, the motif-view leverages a predefined vocabulary of molecular fragments, constructing motifs based on training data insights. The AMDiff joint training paradigm enables information exchange between the two views, integrating geometric and pharmacophoric insights for robust molecular generation.
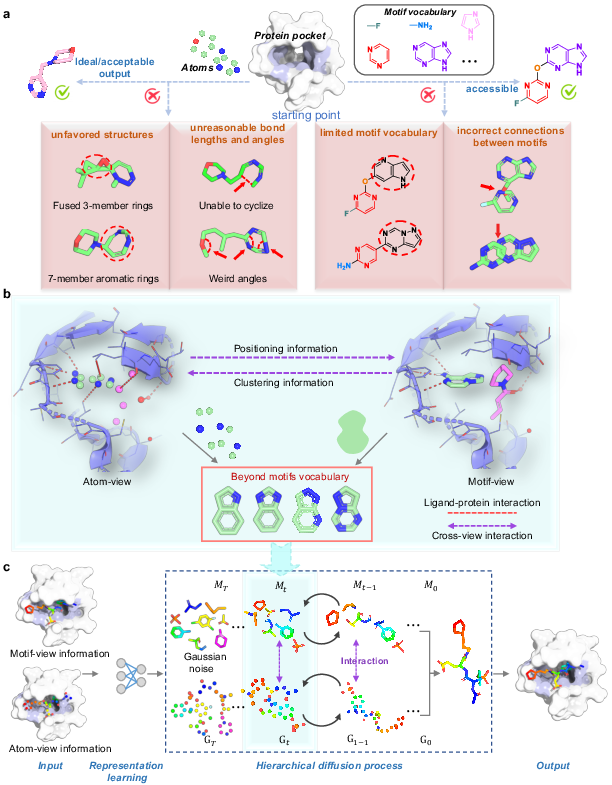
Figure 1: The AMDiff architecture combines atom-view and motif-view through dedicated message-passing networks to leverage signal information in binding site predictions.
Classifier-Free Guidance
AMDiff incorporates a classifier-free guidance approach, enhancing the model's ability to generate desired conditional structures. During the training, the pocket guidance is occasionally omitted to force the model to learn along both the conditional and unconditional pathways. This enables robust generation across various protein targets.
Results and Evaluation
The AMDiff model was evaluated using the CrossDocked dataset and focused on real drug targets such as Anaplastic Lymphoma Kinase (ALK) and Cyclin-dependent kinase 4 (CDK4).
The model achieved superior results in terms of validity, diversity, and novelty. Specifically, it demonstrated a 98.9% validity in generated outputs, highlighting its adherence to chemical bonding rules and realistic conformations.
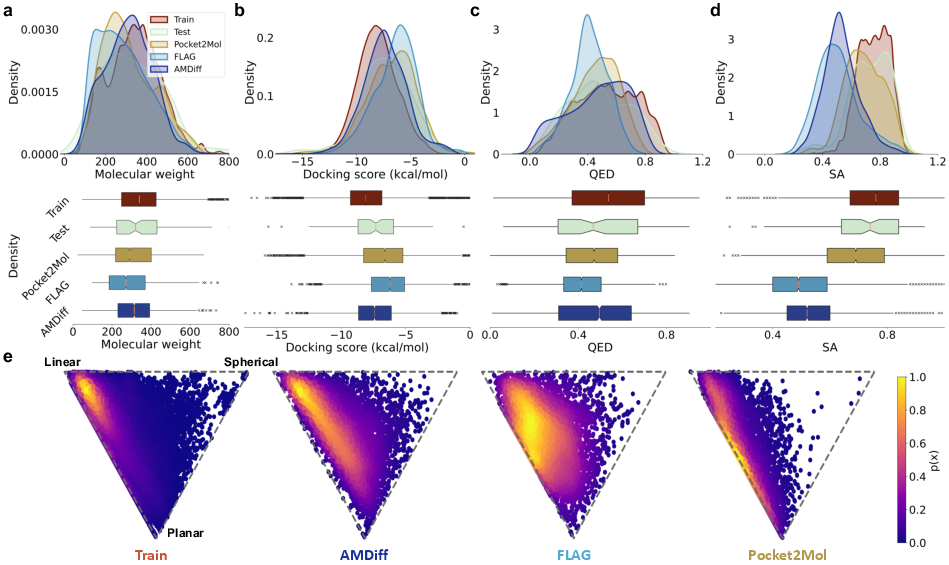
Figure 2: Quantitative performance metrics of AMDiff showing improvements in docking scores and QED, indicating strong interactions with protein pockets.
Generation of Drug-Like Molecules
The AMDiff produced effective molecular structures in real-world therapeutic scenarios, displaying excellent binding affinity and drug-likeness properties. The model showcased its ability to generate molecules with a high degree of interaction and affinity for critical binding sites within the ALK and CDK4 protein complexes.
Topological Features
The integration of topological data analysis metrics, such as persistent homology, enhanced AMDiff's capability to recognize multi-scale geometric and chemical features, offering insights into complex interaction patterns within ligands and proteins.
Discussion
Hierarchical Representation
Building on the importance of a multi-level understanding of biological systems, AMDiff's hierarchical approach aligns with protein structures' inherent organization, offering a powerful tool for de novo drug design.
Limitations and Future Directions
AMDiff does not yet address dynamic conformational changes in proteins, a feature that could further optimize ligand binding efficacy. Incorporating extensive domain-specific features such as pharmacophoric constraints and enhanced validation through laboratory experiments could also refine the model's predictive accuracy.
Conclusion
AMDiff provides an innovative framework for molecule generation, balancing atom-level diversity and motif-level structure with consistency and realism. The model outperforms existing methods, demonstrating significant potential in accelerating the drug discovery process by offering a robust solution to generate effective, novel molecular structures tailored for target protein binding. The integration of hierarchical consistency sets the stage for future advancements in automated drug design within pharmaceutics.

