- The paper introduces novel mathematical models using both deterministic ODEs and stochastic simulations to capture the dynamics of multiple ecDNA types.
- It demonstrates that fitness discrepancies and switching probabilities significantly affect ecDNA distribution and influence tumor evolution.
- The findings highlight the need for tailored cancer therapies targeting ecDNA maintenance and switching mechanisms to combat treatment resistance.
Dynamics of Multiplicity in ecDNA Types
Introduction
The paper "Population dynamics of multiple ecDNA types" (2411.14588) explores the intricate dynamics of extrachromosomal DNA (ecDNA) in cancer cells, presenting novel mathematical models that describe the proliferation and evolution of multiple ecDNA types within a cell population. The study expands on existing models of single ecDNA type dynamics, providing a framework accounting for genetic and phenotypic variations that can influence cancer progression.
Modeling Framework
A core component of this study is its flexible modeling framework (Figure 1), which accommodates different ecDNA types through parameters dictating their interactions.
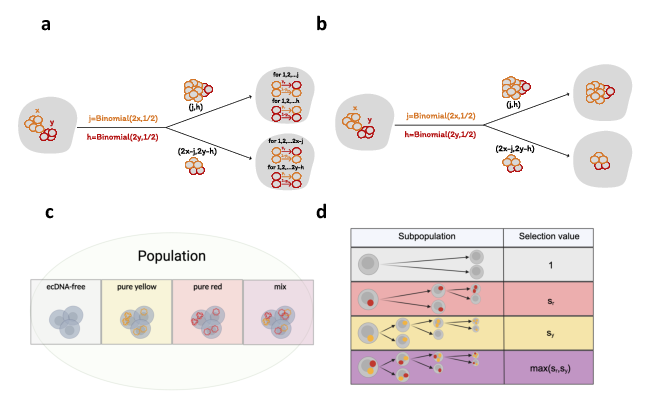
Figure 1: A general framework for modeling two ecDNA types.
The framework allows for modeling distinct species, genotypes with mutations, and phenotypes arising from epigenetic variations or switching. Switching mechanisms are represented via transition probabilities py and pr, illustrating genetic arrangement variabilities or phenotypical changes. The transition probabilities influence whether cells evolve to carry different types of ecDNA, impacting the evolutionary dynamics.
Deterministic and Stochastic Dynamics
Two mathematical methods are used to model ecDNA dynamics: deterministic ordinary differential equations (ODEs) and a stochastic framework (Figure 2). These approaches elucidate the density changes within cellular subpopulations — those carrying ecDNA and those that do not — and how these changes relate to parameters like reproduction rates (sy, sr) and switching probabilities.

Figure 2: ecDNA copy distribution among either identical or non-identical fitness scenarios.
For cells with identical fitness for both ecDNA types, the study finds that type-switching probabilities do not affect ecDNA-positive cells' distribution or ecDNA-free cells' fraction, while fitness discrepancies break this independence.
Analytical and Numerical Insights
The paper discusses analytical solutions aligning with stochastic simulations (Figure 3), particularly focusing on the implications of switching mechanisms on maintaining heterogeneous ecDNA types within a cellular population under different selection scenarios.

Figure 3: Weighted first moment dynamics for ecDNA species.
EcDNA-positive cells maintain different kinetics under identical selective pressures compared to distinct selection strengths. More specifically, one-way and two-way switching impact mean copy numbers differently, evident in the model's results depicting growth dynamics rooted in evolutionary advantages gained through ecDNA's genetic or phenotypic flexibility.
Two-Way Switching's Influence
When examining two-way switching scenarios, the paper's results (Figure 4) suggest that such dynamics encourage a balance between pure and mix cell populations, with switching facilitating greater ecDNA type mixing.

Figure 4: Weighted first moment dynamics in the neutral case for ecDNA geno-/pheno-types, two-way switching.
Switching contributes to sustaining a diverse ecDNA repertoire, thus influencing tumor evolutionary paths and potentially complicating treatment as several ecDNA types may confer differential responses to therapy.
Discussion and Implications
This study provides significant insights into ecDNA dynamics, positing that both genetic mutations and phenotypic state changes significantly influence cancer cell evolution and therapy resistance. The models suggest strategic implications for cancer treatment, underscoring the need for interventions targeting ecDNA maintenance mechanisms and switching pathways to preemptively address potential resistance developments.
Conclusion
The models proposed in this study serve as significant advancements in understanding ecDNA dynamics, incorporating an analytical approach backed by empirical simulation. This framework reveals ecDNA's contribution to cancer's genetic diversity and resistance capabilities, highlighting the necessity of tailored strategies addressing ecDNA-induced heterogeneity in therapeutic contexts.





