- The paper presents Decipher-MR, a model that integrates self-supervised 3D vision learning with report-guided text supervision to generate robust MRI representations.
- It employs a two-stage pretraining strategy combining a 3D ViT-based encoder and a BERT-based text encoder, achieving improved performance in classification, retrieval, segmentation, and localization tasks.
- Key results include a +2.9% AUROC in disease classification and significant improvements in low-data regimes and cross-modal retrieval, demonstrating practical clinical applicability.
Decipher-MR: A Vision-Language Foundation Model for 3D MRI Representations
Introduction and Motivation
The Decipher-MR model addresses the longstanding challenge of developing scalable, generalizable machine learning systems for 3D MRI analysis. MRI's inherent heterogeneity—arising from diverse anatomical coverage, protocol variations, and scanner differences—has historically limited the effectiveness of both traditional and deep learning approaches, especially in low-data or cross-domain settings. While foundation models have demonstrated strong transferability in natural language and 2D vision domains, their application to 3D MRI has been constrained by data scarcity, narrow anatomical focus, and insufficient integration of clinical context.
Decipher-MR is introduced as a 3D vision-language foundation model, pretrained on over 200,000 MRI series from more than 22,000 studies, encompassing a wide range of anatomical regions, imaging protocols, and patient demographics. The model leverages both self-supervised vision learning and report-guided text supervision, aiming to produce robust, generalizable representations that can be efficiently adapted to a broad spectrum of clinical and research tasks via lightweight, modular decoders.
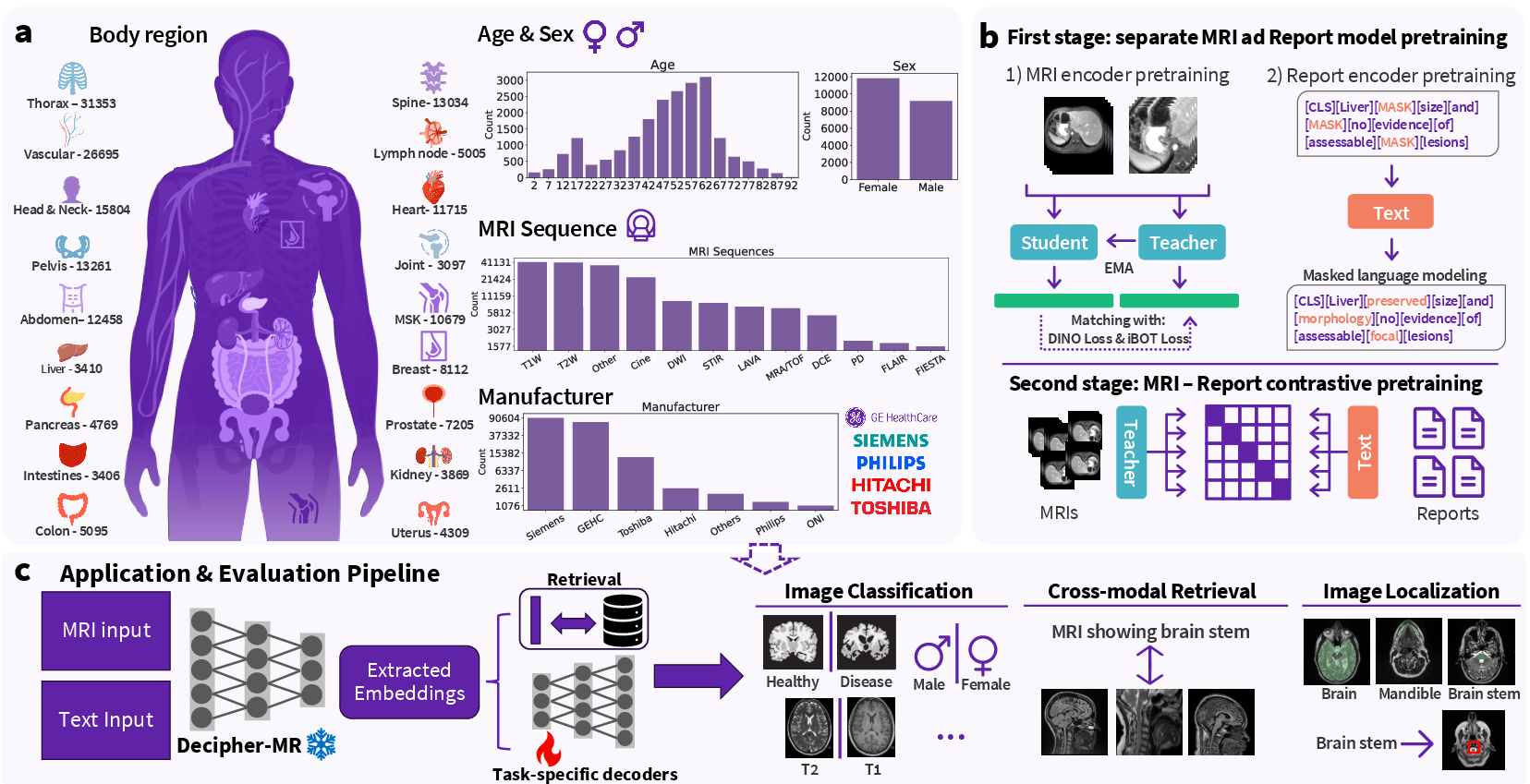
Figure 1: (a) Distribution of the diverse pretraining dataset across age, sex, imaging sequences, body regions, and scanner manufacturers. (b) Overview of the two-stage pretraining framework of Decipher-MR. (c) Evaluation of Decipher-MR in a frozen encoder setup for diverse tasks.
Pretraining Strategy and Model Architecture
Decipher-MR employs a two-stage pretraining pipeline:
- Stage 1: Unimodal Self-Supervised Pretraining
- Vision Encoder: A 3D ViT (ViT-Base, 86M parameters) is pretrained using a DINOv2-based student-teacher framework, combining masked image modeling and multi-crop contrastive objectives. The model is robust to variable input sizes via trilinear position embedding interpolation and is trained with extensive data augmentation to handle MRI-specific inconsistencies.
- Text Encoder: A BERT-based model (initialized from PubMedBERT) is pretrained on radiology reports using masked LLMing, capturing domain-specific linguistic patterns.
- Stage 2: Vision-Language Contrastive Alignment
- The pretrained vision and text encoders are further aligned using image-report contrastive learning, mapping both modalities into a shared 512-dimensional embedding space. Organ-based batch sampling and report section extraction (via LLMs and SNOMED mapping) are used to enhance fine-grained anatomical and pathological alignment.
This modular design enables the pretrained encoder to remain frozen during downstream adaptation, with task-specific decoders (e.g., MLPs, ResNet-based segmentation heads, transformer-based localization modules) trained atop the fixed backbone.
Evaluation Across Downstream Tasks
Classification and Regression
Decipher-MR demonstrates consistent improvements over state-of-the-art baselines (DINOv2, BiomedCLIP, MedImageInsight, SAMMed3D) on a range of classification and regression tasks, including disease diagnosis, demographic prediction, body region identification, and imaging attribute detection. Notably, the model achieves:
- +2.9% AUROC in disease classification
- +3.0% in demographic prediction
- Superior performance in low-data regimes, with the performance gap widening as labeled data decreases
Bias analysis reveals that Decipher-MR maintains higher robustness across sex-based splits, outperforming MedImageInsight by an average of 5.5% in cross-sex evaluation scenarios.
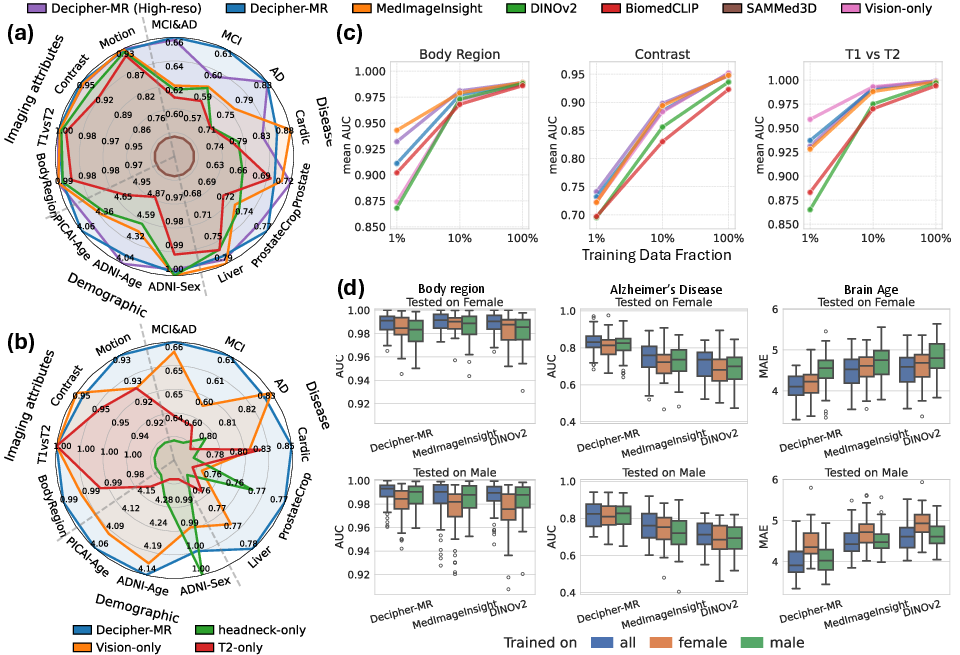
Figure 2: (a) Classification performance comparison using MLP probes. (b) Ablation of pretraining data diversity and text supervision. (c) Performance under low-data regimes.
Ablation studies confirm that both anatomical/protocol diversity and text supervision are critical for generalization. Models pretrained on restricted anatomical regions or protocol types underperform even on in-domain tasks, and the addition of report-guided contrastive learning yields significant gains, especially in cardiac and prostate lesion classification, brain age regression, and body region detection.
Cross-Modal Retrieval
Decipher-MR's vision-language alignment enables zero-shot cross-modal retrieval, supporting both text-to-image and image-to-text queries. On out-of-domain datasets:
- Body region retrieval (Source1): Top-3 localization success rate of 91.4% (full reports) and 78.8% (short descriptions), outperforming MedImageInsight by 10–15%.
- Tumor pathology retrieval (Source2): 39.3% accuracy in retrieving correct tumor subregion descriptions, surpassing BiomedCLIP and MedImageInsight by 12–20%.
The model achieves higher mean average precision (mAP) and greater retrieval diversity, particularly when using concise, clinically relevant queries.
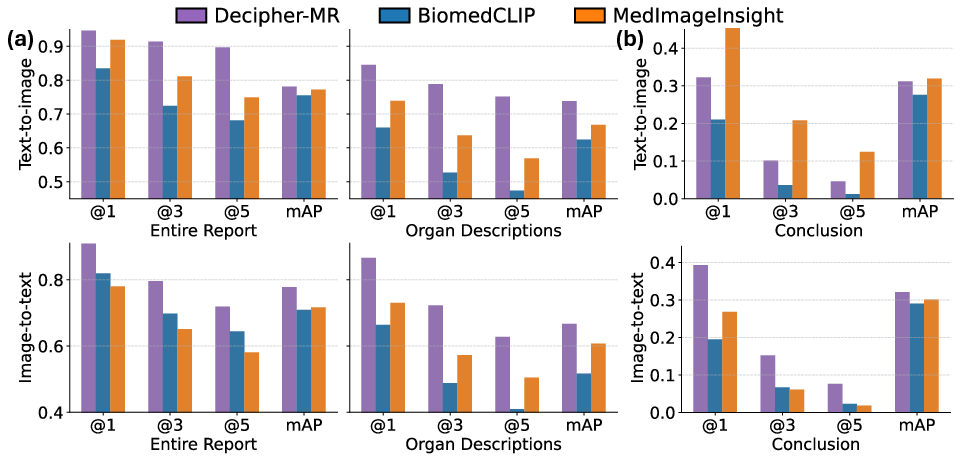
Figure 3: Cross-modal retrieval performance for body region and tumor pathology tasks.
Segmentation and Localization
For 3D anatomical segmentation, Decipher-MR (frozen encoder + ResNet decoder) matches or exceeds nnUNet (fully end-to-end) and outperforms MedSAM3D (frozen encoder) across abdominal, pelvic, and cardiac datasets. The model converges rapidly, achieving high Dice scores within a single epoch, and demonstrates strong performance even with minimal fine-tuning.
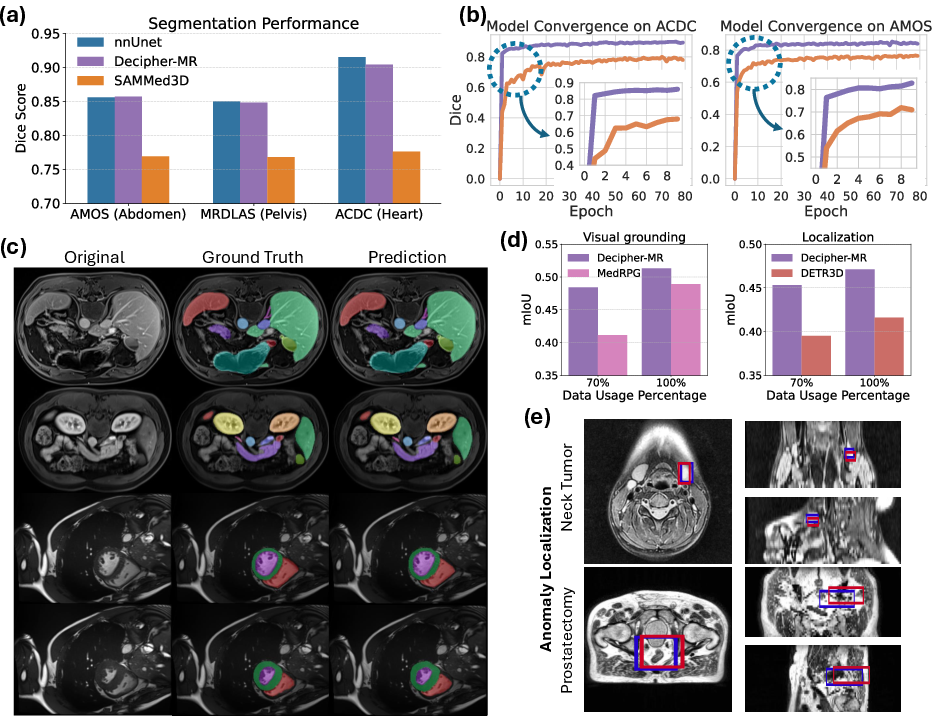
Figure 4: (a) Segmentation performance across datasets. (b) Convergence speed comparison. (c) Qualitative segmentation examples. (d) Anomaly localization and visual grounding. (e) Qualitative anomaly localization results.
For anomaly localization and visual grounding, Decipher-MR as a frozen encoder yields 14% higher mIoU in localization and 11.5% higher mIoU in visual grounding compared to end-to-end trained DETR3D and MedRPG models, respectively. Qualitative results show accurate detection of tumors and surgically removed organs across diverse anatomical regions.
Implementation Considerations
- Data Processing: Foreground cropping (Otsu's method), intensity normalization, and resolution standardization are essential for robust pretraining. Report processing leverages LLMs for section extraction and SNOMED mapping for anatomical consistency.
- Modular Adaptation: The frozen encoder paradigm enables rapid adaptation to new tasks with minimal computational overhead, supporting efficient development pipelines in clinical settings.
- Preprocessing Trade-offs: Task-specific preprocessing (e.g., cropping, resizing, skull-stripping) can further boost performance but may reduce generalizability for tasks requiring broader context.
- Resource Requirements: Pretraining is computationally intensive (100+ epochs, large batch sizes), but downstream adaptation is lightweight due to the frozen encoder approach.
Limitations and Future Directions
- Textual Supervision: Gains from report-guided pretraining are less pronounced for imaging attribute classification; richer DICOM metadata and more diverse report corpora may be needed.
- Pathology-Focused Retrieval: Performance in fine-grained pathology retrieval is limited by the diversity and granularity of textual data.
- Bias and Generalization: While Decipher-MR is more robust to demographic variation, performance drops persist in cross-sex and out-of-distribution scenarios, indicating the need for further bias mitigation and domain adaptation strategies.
- Encoder Fine-Tuning: While the frozen encoder paradigm is efficient, certain applications may benefit from partial or full encoder fine-tuning, especially for highly specialized tasks.
Conclusion
Decipher-MR establishes a new standard for MRI-specific foundation models by integrating large-scale, diverse 3D MRI data with report-guided vision-language pretraining. The model demonstrates strong, generalizable performance across classification, retrieval, segmentation, and localization tasks, often matching or exceeding specialized end-to-end models with significantly reduced adaptation cost. Its modular, frozen-encoder design supports efficient, scalable development of AI solutions for MRI, with broad implications for clinical and research applications. Future work should focus on expanding textual supervision, enhancing pathology-specific retrieval, and developing advanced bias mitigation and fine-tuning strategies to further improve generalizability and fairness.





