- The paper introduces a multi-modal flow matching approach that combines continuous methods for atomic coordinates and discrete techniques for categorical features to generate 3D molecules.
- It leverages innovations like self-conditioning, fake atoms, and geometry distortion to enhance model stability and ensure accurate geometric reconstruction.
- Experimental results demonstrate nearly 100% molecular validity and superior functional group fidelity compared to baseline models, underscoring its efficiency and scalability.
FlowMol3: Flow Matching for 3D De Novo Small-Molecule Generation
FlowMol3 introduces an innovative approach to de novo molecule generation using a multi-modal flow matching framework. It aims to improve on previous methods for generating small drug-like molecules in three dimensions by employing architecture-agnostic enhancements. This essay discusses the paper's methodology, implementation details, and real-world applications, providing insights into how FlowMol3's features can be harnessed in practical scenarios.
Introduction to FlowMol3
FlowMol3 achieves notable progress in the generative modeling of small molecules by leveraging a unique multi-modal flow matching approach. This method involves sampling atomic coordinates through continuous flow matching while using discrete flow matching for atom types, charges, and bond orders. Key innovations include the introduction of self-conditioning, fake atoms, and geometry distortion techniques that improve model stability and output quality, without increasing computational costs.
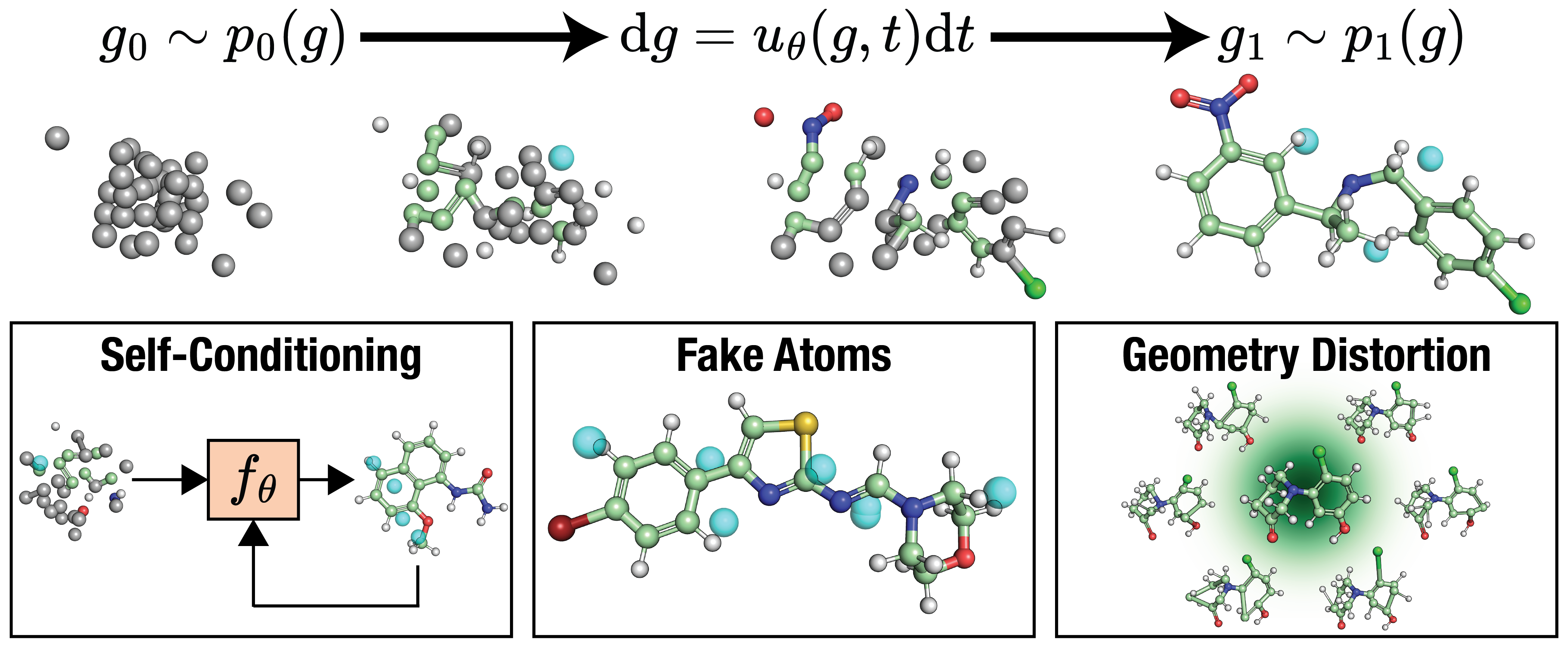
Figure 1: FlowMol3 Overview: FlowMol3 is a multi-modal flow matching model for unconditional de novo molecule generation. Atomic coordinates are generated by sampling an ordinary differential equation learned via continuous flow matching. Atom types, charges, and bond orders are generated by simultaneous simulation of Continuous-Time Markov Chains (CTMCs) learned by discrete flow matching. We find three features to be critical for improved model performance: self-conditioning, fake atoms, and geometry distortion.
Model Design and Implementation
Continuous and Discrete Flow Matching
FlowMol3 is designed to cope with both continuous and categorical data modalities inherent in molecular graphs:
- Continuous Flow Matching (CFM): Utilized to predict atomic coordinates efficiently via deterministic interpolants and vector fields. The conditional probability paths are formulated as lines connecting initial and terminal atomic states, allowing for precise spatial positioning during molecule generation.
- Discrete Flow Matching (DFM): Implemented using Continuous-Time Markov Chains (CTMCs) to manage categorical variables like atom types and bond orders. This approach ensures the smooth transformation between categorical states over time, maintaining molecular identity accuracy.
Enhancements for Robustness
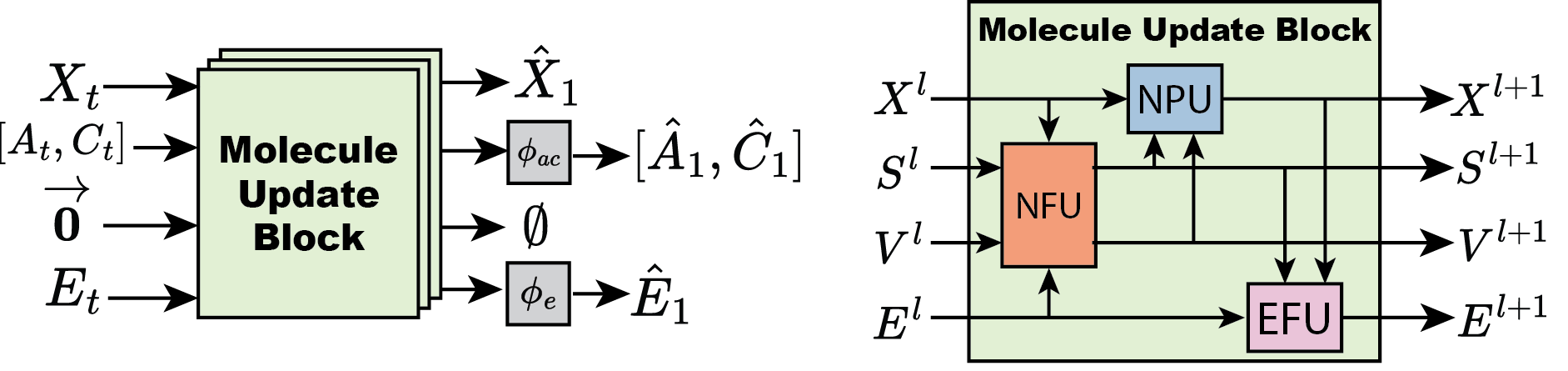
Figure 2: FlowMol Architecture Left: An input molecular graph is transformed into a predicted final molecular graph by being passed through multiple molecule update blocks. Right: A molecule update block uses NFU, NPU, and EFU sub-components to update all molecular features.
FlowMol3 integrates several architecture-agnostic innovations:
- Self-Conditioning: This mechanism allows the model to refine its predictions iteratively, using its past outputs as inputs for subsequent iterations, thereby increasing accuracy.
- Fake Atoms: Introduced as a mode to adjust the number of atoms in the system dynamically, improving adaptability to unexpected geometric configurations.
- Geometry Distortion: Applied during training to expose the model to perturbed atomic structures, enhancing its capacity to recover from off-distribution states during inference.
Experimental Evaluation
FlowMol3 surpasses previous models across various metrics for 3D molecular generation. It demonstrates nearly 100% validity for generated molecules. Furthermore, it effectively reproduces the functional group composition and geometric configurations seen in its training data, achieving this with significantly fewer learnable parameters compared to its contemporaries.
Comparisons with Baseline Models
FlowMol3 shows superior performance against baseline models like SemlaFlow, Megalodon, and ADiT in terms of molecular validity and functional group fidelity. The model's parameter efficiency and enhanced sampling speed without sacrificing quality underscore its practical applicability in computational chemistry.
Practical Implications and Future Work
FlowMol3's innovations have direct implications for accelerating computational drug discovery by enhancing the fidelity and utility of generated molecular structures. Its efficient and scalable framework lays groundwork for broad applications beyond molecule design, extending into areas like protein simulation and material science.
Future exploration may focus on integrating FlowMol3's enhancements into conditional generation frameworks or further theoretical work to tighten the bounds on distribution drift. These directions promise to extend the model's applicability while refining its robustness and precision.
Conclusion
FlowMol3 represents a significant advance in the generative modeling of small molecules, employing a robust, efficient, and architecture-agnostic strategy that achieves outstanding molecular generation fidelity. Its features are transferable and could catalyze further improvements in both the science of molecular design and the broader field of AI-driven material discovery.

