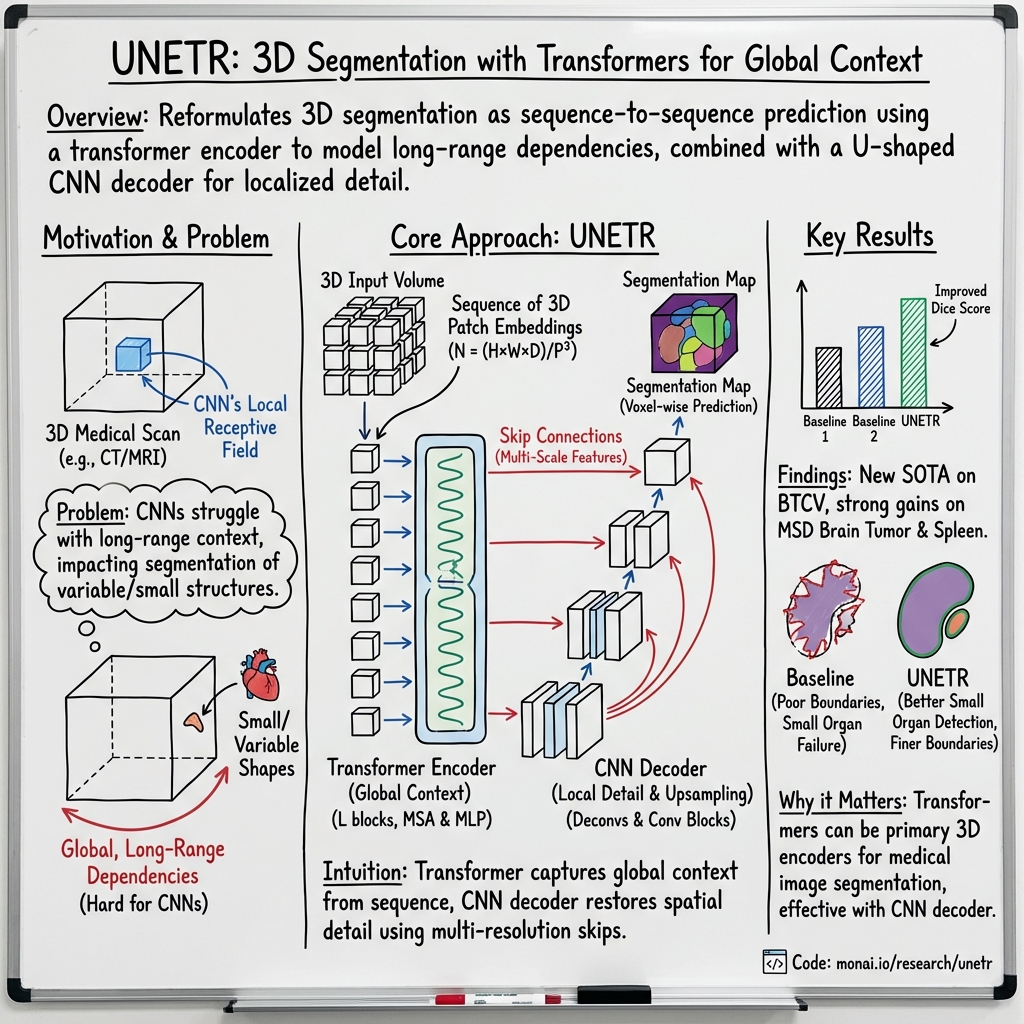
- The paper introduces UNETR, a novel transformer-based architecture that reimagines 3D medical image segmentation as sequence-to-sequence prediction.
- The paper integrates a transformer encoder with a CNN-based decoder via skip connections, effectively fusing global context and local details.
- The paper demonstrates state-of-the-art performance on BTCV and MSD datasets, achieving higher Dice scores especially in segmenting small and complex anatomical structures.
The paper introduces a novel approach for 3D medical image segmentation using a Transformer-based architecture, denoted as UNEt TRansformers (UNETR). This methodology leverages the long-range dependency learning capabilities of transformers, reimagining the segmentation task as a sequence-to-sequence prediction.
Introduction and Motivation
Fully Convolutional Neural Networks (FCNNs) have been preeminent in medical image segmentation due to their ability to learn both global and local features via U-Net-like encoder-decoder architectures. However, the intrinsic limitation of convolutional layers in FCNNs is their localized receptive field, which constrains their capacity to capture long-range spatial dependencies. This limitation often results in suboptimal segmentation, particularly of structures with variable shapes and scales.
The transformer model, which has achieved significant success in NLP by learning long-range dependencies through self-attention mechanisms, presents an opportunity to address this shortcoming in image segmentation tasks. The paper extends this transformative approach to volumetric (3D) medical image segmentation, proposing UNETR to harness the power of transformers for learning global context in medical images.
Methodology
UNETR deploys a transformer as the encoder within a U-Net-like architecture, directly processing 3D patches of the input volume. It preserves the benefits of self-attention modules to model long-range dependencies and capture global context. The architecture is distinct for its direct linkage of transformer encoder outputs to a CNN-based decoder via skip connections, facilitating multi-scale information fusion for accurate segmentation outputs.
Architecture Details
The architecture comprises:
- A transformer encoder that operates on a 1D sequence of embeddings derived from 3D input volume patches.
- Skip connections from the transformer encoder to the CNN-based decoder at multiple resolutions.
- Application of deconvolutional layers at the encoder's bottleneck to increase feature resolution, followed by concatenation with previous transformer outputs for hierarchical multi-scale information integration.
Experimental Validation
UNETR's efficacy was validated on the BTCV dataset for multi-organ segmentation and the MSD dataset for brain tumor and spleen segmentation. Benchmarks demonstrate that UNETR surpasses state-of-the-art performance in both datasets.
BTCV Dataset
For the BTCV dataset, UNETR achieved new state-of-the-art performance in both the Standard and Free Competitions sections. Specifically, in the Free Competition, UNETR obtained an average Dice score of 0.899, significantly outperforming competing models by substantial margins in both average Dice score across different anatomical structures and segmentation accuracy, especially for small organs.
MSD Dataset
In the MSD dataset, UNETR demonstrated superior segmentation performance for brain tumors and spleen segmentation. For brain tumor segmentation, it excelled in segmenting subregions like the tumor core, whole tumor, and enhancing tumor core. In spleen segmentation, UNETR consistently outperformed other models by achieving higher Dice scores.
Qualitative Analysis
Qualitative assessments further highlight UNETR's precise delineation of organ boundaries and its competency in segmenting low-contrast tissues adjacent to other anatomical structures. This qualitative evaluation underscores the advantage of incorporating long-range dependency learning through transformers.
Discussion and Implications
UNETR's deployment reveals significant performance advantages by incorporating global contextual information through transformers into the segmentation pipeline. This effectively addresses the main limitation of traditional FCNNs, enhancing performance in segmenting complex and small-scale anatomical structures.
The study opens avenues for further exploration in AI-driven medical imaging:
- Future iterations could focus on optimizing transformer components to balance computational complexity and segmentation accuracy.
- Investigations into pre-trained transformer models tailored for medical image datasets could further elevate performance.
- Extending UNETR to other medical imaging tasks, including 3D object detection and volumetric reconstruction, could reveal wider applicability.
Conclusion
UNETR sets a new benchmark for 3D medical image segmentation by integrating transformers to capture long-range dependencies. The architecture’s capacity to effectively merge global and local information represents a significant advancement in medical image analysis. This foundation offers a promising direction for subsequent research into transformer-based models in the field.