- The paper introduces ClinicalGPT-R1, employing diversified real-world and synthetic data with a two-stage training process (SFT and RL) to improve clinical reasoning.
- It demonstrates enhanced diagnostic performance by utilizing sophisticated data synthesis and evaluation benchmarks over models like GPT-4o and Qwen2.5-7B-Instruct.
- The study highlights the impact of language-specific datasets and advanced methodologies in elevating diagnostic accuracy in clinical settings.
Introduction
The paper "ClinicalGPT-R1: Pushing reasoning capability of generalist disease diagnosis with LLM" explores the application of LLMs in medical diagnostics, focusing on enhancing the reasoning capabilities essential for accurate disease diagnosis. The authors introduce ClinicalGPT-R1, a model specifically designed to address challenges in clinical reasoning through the use of rich datasets and sophisticated training strategies.
Methods
Data Collection and Synthesis
ClinicalGPT-R1 was trained using a diversified dataset comprising real-world clinical records and synthetic data. The real-world data was sourced from MedDX-FT and included anonymized electronic health records (EHRs). Synthetic data creation involved state-of-the-art LLMs utilizing a sophisticated pipeline to simulate diagnostic reasoning processes. The pipeline emphasized accurate final outcomes and comprehensive reasoning strategies, including techniques like Exploring New Paths, Backtracking, and Corrections.

Figure 1: The pipeline of synthesized data utilized in training ClinicalGPT-R1.
Training Strategies
Training ClinicalGPT-R1 involved a two-stage process: Supervised Fine-Tuning (SFT) and Reinforcement Learning (RL). Initially, SFT was applied to instill a reasoning-first approach in the model, focusing on explicit reasoning steps prior to decision-making. Then, RL further optimized these reasoning trajectories using Proximal Policy Optimization (PPO), enhancing the decision-making quality based on a structured reward system.
Evaluation
For evaluation, the authors introduced MedBench-Hard, a benchmark comprising diverse diagnostic cases spanning seven medical specialties. ClinicalGPT-R1 demonstrated superior reasoning and diagnostic capabilities compared to other models such as GPT-4o and Qwen2.5-7B-Instruct, particularly in Chinese language tasks.
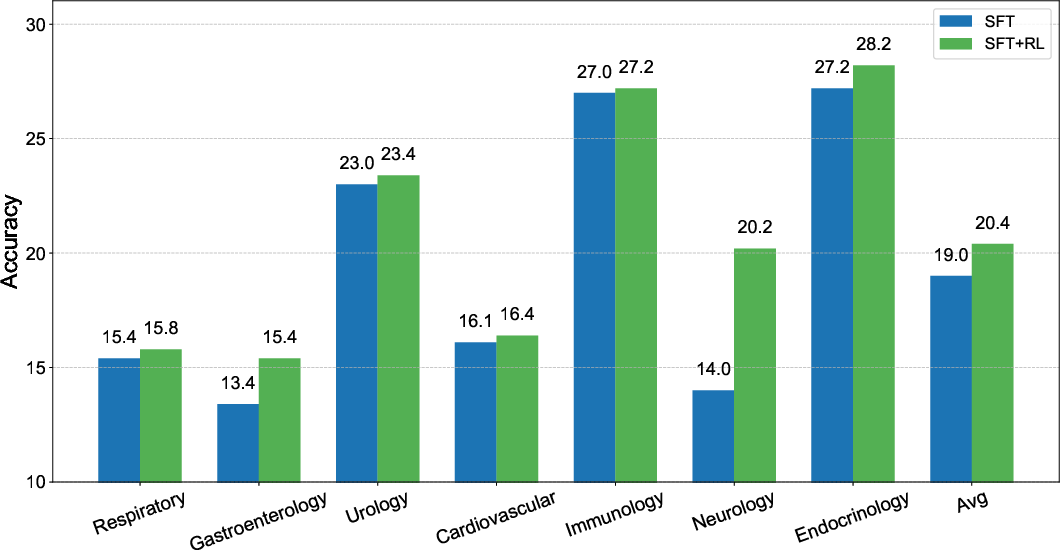
Figure 2: Comparison of ClinicalGPT-R1 with different training methods, illustrating benefits of the combined SFT and RL approach.
Language and Data Source Influence
Experimental results indicated significant performance variations based on the language and data sources. Models trained on datasets synthesized by GPT-4o-mini outperformed those trained with Deepseek-v3-0324-generated data. Moreover, the Chinese language datasets yielded more robust diagnostic performance compared to English datasets.
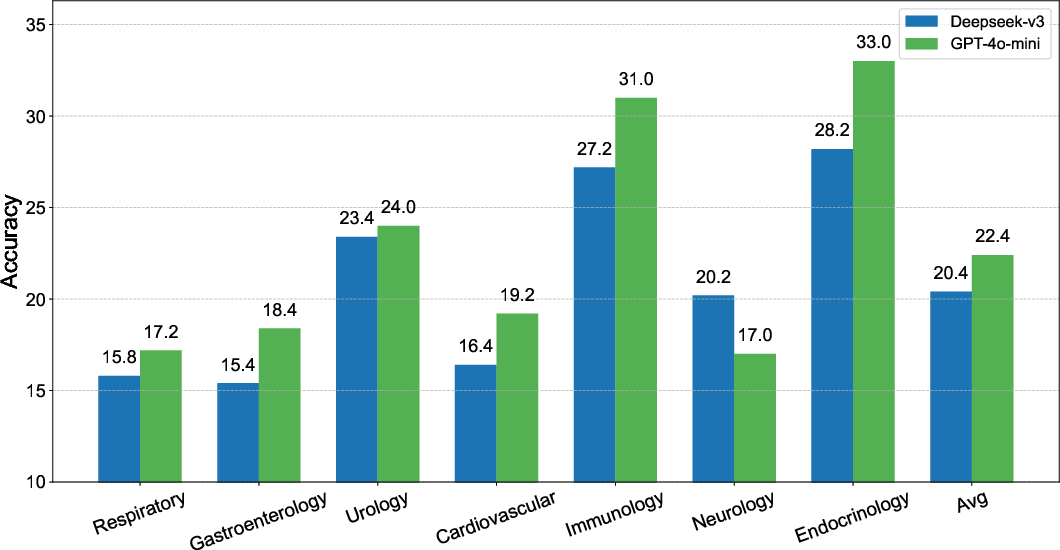
Figure 3: Comparison of ClinicalGPT-R1 with different training data sources highlighting source influence.

Figure 4: Effect of language on model performance showing superior results with Chinese datasets.
The comparison between ClinicalGPT-R1 and baseline models further validated the former's strength across multilingual environments. In Chinese diagnostic tasks, ClinicalGPT-R1 achieved superior performance, while in English, it matched GPT-4o's performance and surpassed Qwen2.5-7B-Instruct.

Figure 5: Model performance in Chinese language tasks, showcasing ClinicalGPT-R1's superiority.

Figure 6: Performance of ClinicalGPT-R1 in English tasks compared to baselines, affirming competitive diagnostic capability.
Conclusion
ClinicalGPT-R1 represents a significant advancement in LLM application for clinical diagnostics, particularly through enhanced reasoning capabilities critical for disease diagnosis. This study's findings underscore the importance of diverse training strategies and high-quality data synthesis in optimizing LLMs for complex medical reasoning tasks. Future explorations might include expanding this approach to other medical specialties and further refining language-specific adaptations to maximize diagnostic accuracy.





