- The paper introduces a 3D U-Net model that uses dense 3DRA annotations to significantly improve segmentation accuracy for bAVM vascular structures.
- Methodology incorporates self-configuring parameters, deep supervision, and combined loss functions to effectively capture complex vascular morphologies.
- Results show enhanced Dice and Recall metrics along with superior qualitative vessel coverage compared to traditional thresholding and region growing methods.
Deep Learning for Brain Vessel Segmentation in 3DRA with bAVMs
This paper introduces a 3D U-Net-based deep learning model for segmenting brain vessels in 3D rotational angiography (3DRA) images of patients with brain arteriovenous malformations (bAVMs). The model addresses the challenges of bAVM segmentation, including the difficulty in obtaining annotated data and the complex vascular structures associated with bAVMs. The paper demonstrates that the proposed deep learning approach outperforms traditional methods, achieving a comprehensive coverage of relevant structures for bAVM analysis.
Methodology and Architecture
The authors densely annotated 5 3DRA volumes of bAVM cases to train and evaluate their model. The dataset includes feeding arteries, draining veins, AVM nidus, and fistulas. The 3D U-Net architecture (Figure 1) is based on the nnUNet framework, adapting to the dataset's characteristics through self-configuration of parameters such as the number of layers, convolutions, patch size, and batch size. The network uses patches of 64×64×64 pixels, downsampling via strided convolutions in the encoder and upsampling with transposed convolutions in the decoder. Instance normalization and leaky ReLU are applied after each convolutional layer, and deep supervision is incorporated using additional segmentation outputs from the last four decoder layers.
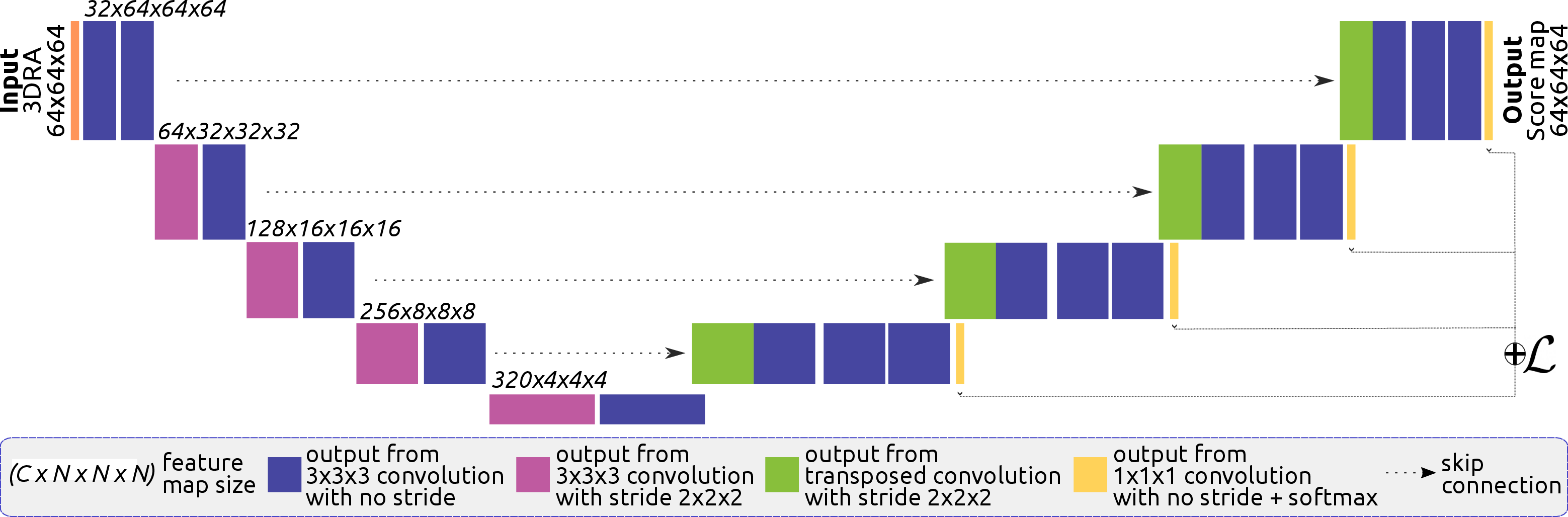
Figure 1: Architecture of the proposed 3DUNet for brain vessel segmentation in bAVM cases.
The model employs a leave-one-out cross-validation strategy (Figure 2), training five models with a batch size of 2, an initial learning rate of 0.01, and stochastic gradient descent with Nesterov momentum for 250 epochs. Two loss functions were compared: a combined cross-entropy and Dice loss (Lcombo) and a combined cross-entropy, Dice, and soft centerline Dice loss (Lcombo+clDice).
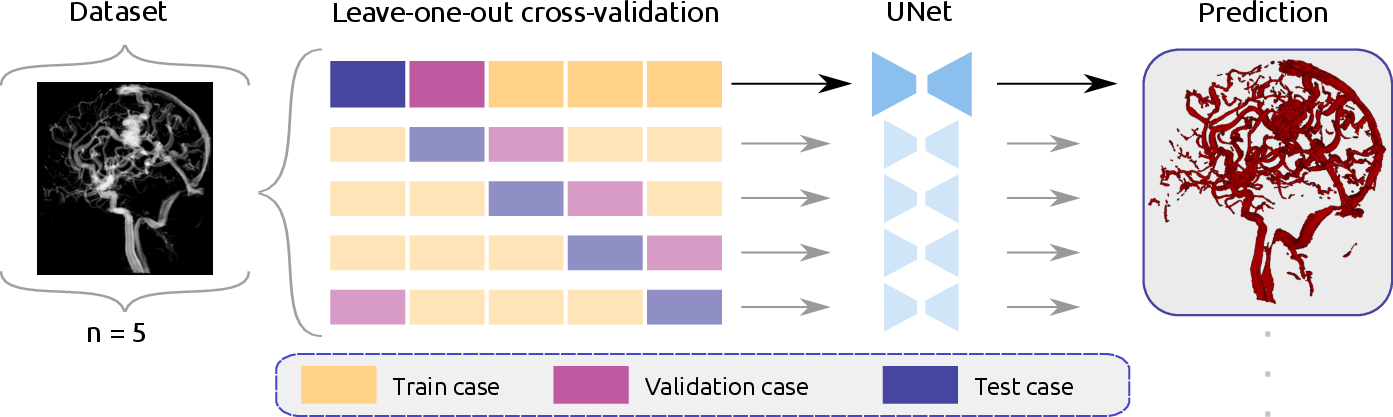
Figure 2: Schematic overview of the leave-one-out cross-validation training.
Results and Evaluation
The models were evaluated quantitatively using Dice, Recall, and Precision metrics for both overall vessels and centerline approximations. Qualitative assessments were also conducted to visually inspect the segmentation results. Quantitative results demonstrated that the 3DUNet models outperformed traditional thresholding and region growing methods. Specifically, the UNet models achieved higher Dice and Recall values, indicating better coverage of foreground voxels. While region growing exhibited higher Precision values, its overall recovery of foreground voxels was insufficient.
Qualitatively, the 3DUNet models demonstrated a higher discovery rate of overall vessel structures, including draining vessels and venous structures often missed by thresholding and region growing. The models also identified distal arteries and structures not present in the ground truth, highlighting the ability to capture finer vascular details (Figures 3 and 4).
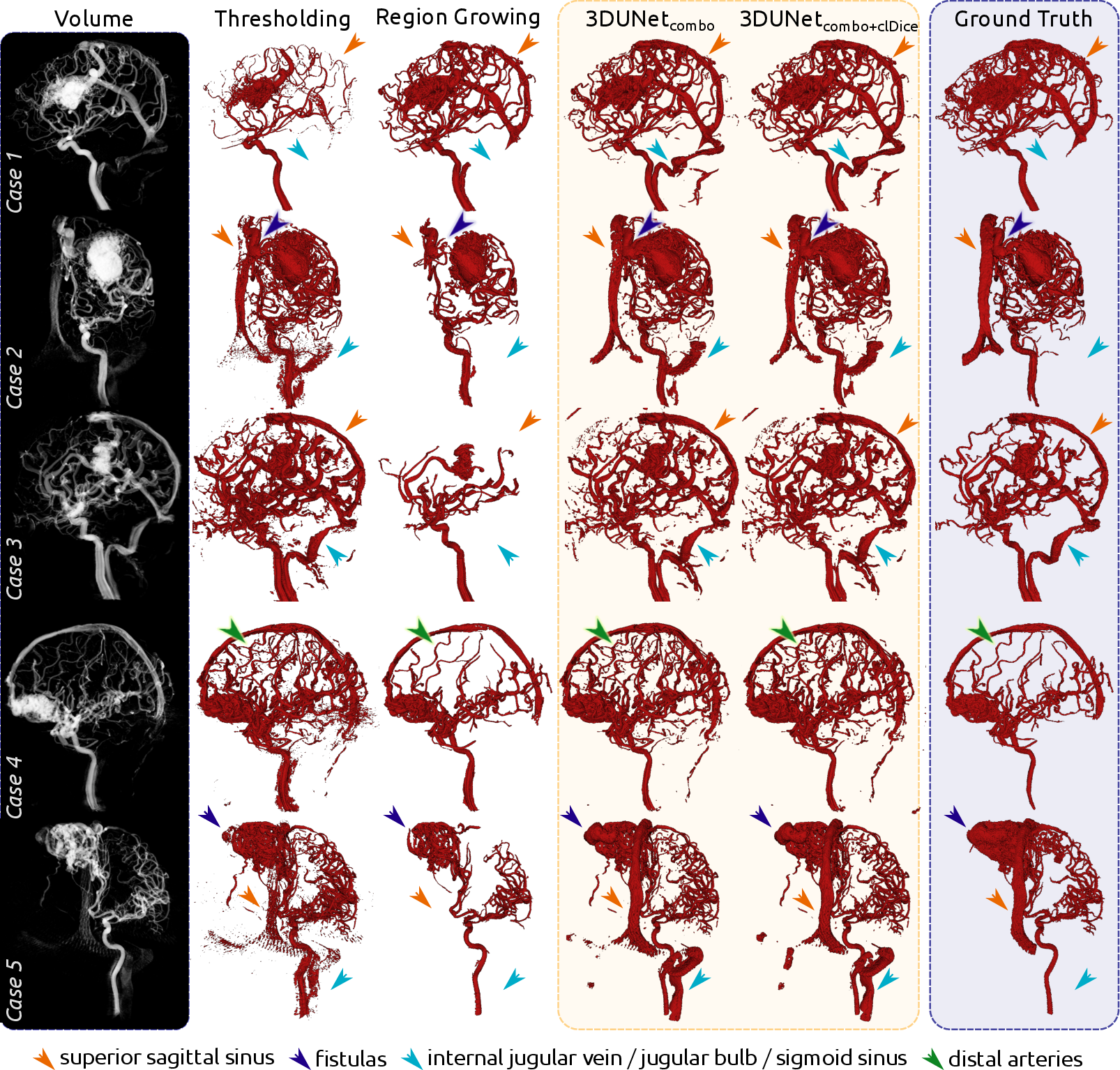
Figure 3: Qualitative results for each of the five cases, comparing volume rendering, segmentations, and ground truth.
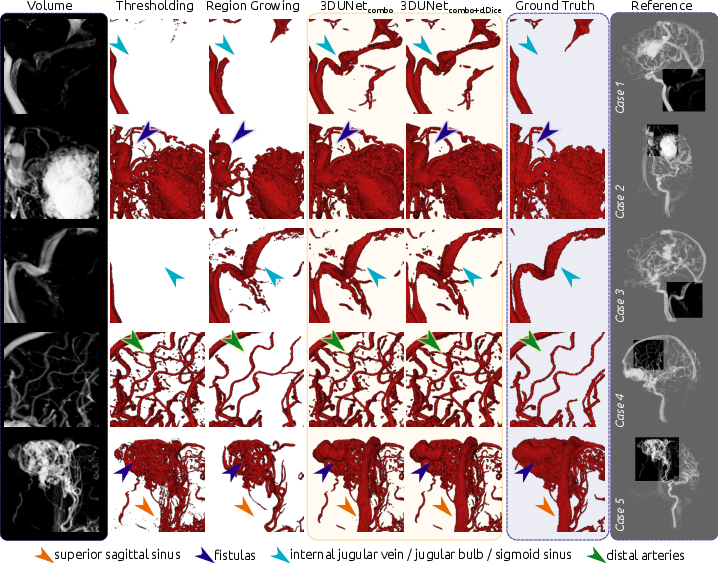
Figure 4: Enlarged view of the qualitative results, highlighting vein structures, fistulas, distal arteries, and dense nidus.
Discussion and Implications
The paper addresses the critical issue of bAVM segmentation in 3DRA images, offering a deep learning solution that mitigates the challenges of data scarcity and complex vascular morphologies. The use of a centerline-focused loss function provides enhanced connectivity and clearer vessel delineations. The false positives identified by the models, primarily veins and distal arteries not delineated in the ground truth, suggest the potential to improve manual annotation processes by providing more comprehensive initial segmentation guesses. Furthermore, the model's ability to segment venous structures is valuable for transvenous embolization planning. Future work may focus on the inclusion of data augmentation techniques and synthetic vessel datasets to further improve model robustness.
Conclusion
This work presents a promising deep learning approach for segmenting bAVMs in 3DRA images, addressing the limitations of traditional methods and the challenges of manual annotation. The proposed model achieves a satisfactory coverage of relevant structures, potentially alleviating the burden of manual labeling and serving as a starting point for more reliable segmentations, ultimately contributing to a better understanding and characterization of bAVMs.



