- The paper introduces a novel 3D VQ-VAE model that compresses brain MRI data to under 1% while maintaining structural integrity.
- It employs innovative 3D convolutional architectures and adaptive robust loss to overcome challenges typical in high-dimensional training.
- Experimental results show significant improvements in MMD, MS-SSIM, and Dice scores compared to traditional GAN and VAE approaches.
Neuromorphologicaly-preserving Volumetric Data Encoding using VQ-VAE
This paper presents a novel application of Variational Autoencoders (VQ-VAE) for encoding high-dimensional medical volumetric data. Specifically, it demonstrates the capability to compress 3D brain MRI data to less than 1% of its original size while preserving morphological integrity, thereby achieving significant improvements over prior methodologies.
Background and Motivation
The task of encoding high-resolution 3D volumetric medical data presents unique challenges due to the intricate and high-dimensional nature of such data. Traditional approaches, such as GANs and VAEs, have exhibited limitations in terms of fidelity and morphological preservation, particularly within the context of medical imaging where structural precision is paramount. This paper leverages the advantages of VQ-VAE, initially developed to mitigate GAN limitations, by substituting Gaussian priors with a vector quantization process and aligning it to capture volumetric characteristics effectively.
Model Architecture
The architectural adaptation of VQ-VAE for 3D inputs forms the core contribution of this work. The network's innovative design incorporates 3D convolutions and effective quantization strategies, as depicted in the complex hierarchical network structure.
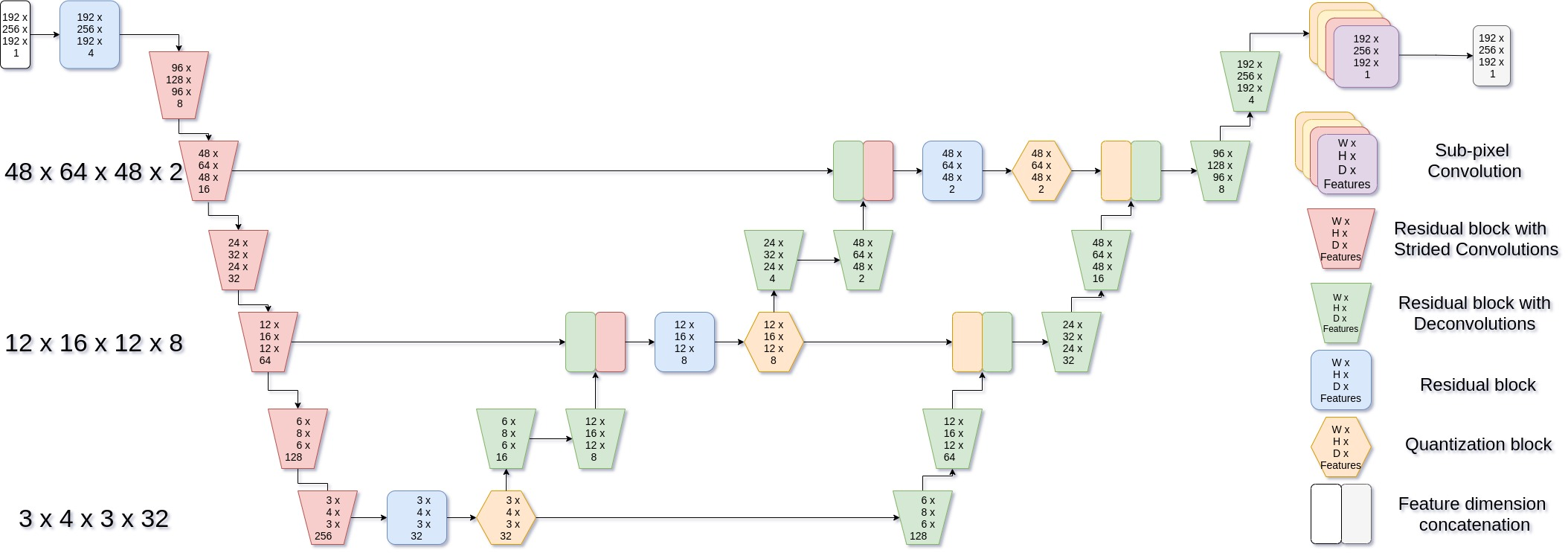
Figure 1: Network architecture - 3D VQ-VAE.
By integrating SubPixel Deconvolutions and FixUp blocks, the model adeptly overcomes common constraining artifacts and stabilization issues typically encountered in high-dimensional training scenarios.
Loss Functions and Training Procedures
The model employs enhanced loss functions to ensure high fidelity. Among these, the paper explores an adaptive robust loss, allowing the model to adjust dynamically throughout training to optimize dimensional representation beyond raw data metrics.
Additionally, the paper retains classical VQ-VAE codebook loss strategies, including updates via Exponential Moving Average (EMA), fostering stable and fast convergence. These modifications significantly bolster the model's capacity to preserve critical neuromorphological structures.
Datasets and Experimental Setup
The model's efficacy was validated using datasets from ADNI, OASIS, and additional neurologically diverse imaging sources. Preprocessing steps involved precise brain extraction methods and registration techniques to ensure uniform and robust inputs across experiments. Metrics such as maximum mean discrepancy (MMD) and multi-scale structural similarity (MS-SSIM) were pivotal in assessing reconstruction fidelity.
Experimental Results
The model notably surpasses prior work in reconstruction metrics, achieving MMD and MS-SSIM enhancements critical for clinical applicability.
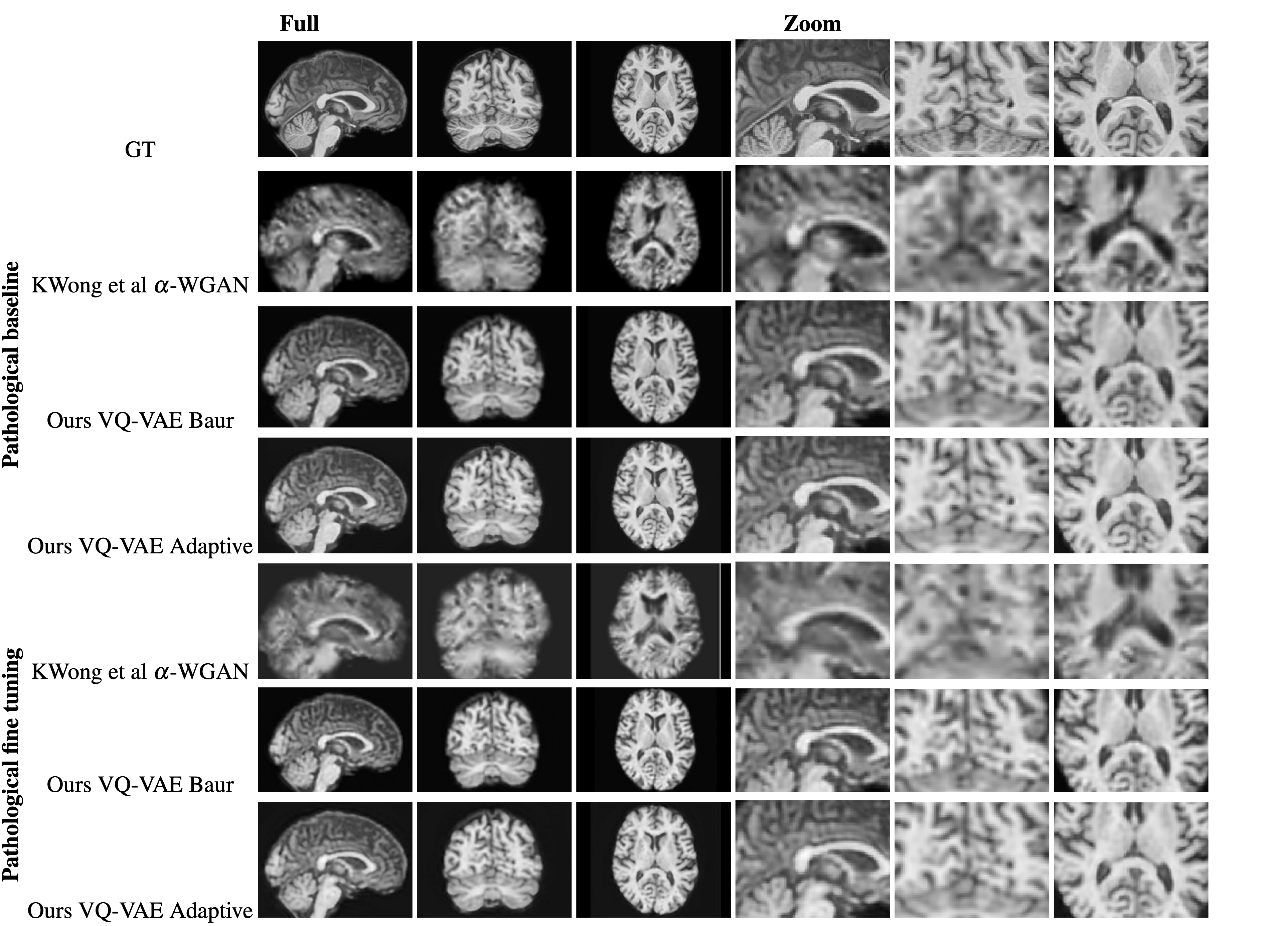
Figure 2: Comparison of a random reconstruction across the pathological training modes. Alpha-WGAN's reconstruction was upsized to 1mm isotropic.
Achieving exceptional Dice scores for white matter, gray matter, and CSF segmentations substantiates the model's precision in maintaining anatomical structural integrity.
Further validation is undertaken through Voxel-Based Morphometry (VBM) analyses evaluating reconstruction impacts and illustrating significant improvements in morphological preservation over traditional GAN-based approaches.

Figure 3: VBM Two-sided T-test between AD images and their reconstructions.
Conclusion
The proposed VQ-VAE model exemplifies a significant leap in neuromorphologically preserving volumetric data encoding. Its methodology balances compression with meticulous structural fidelity, promising advancements in medical imaging fields. Future exploration could enhance fine-tuning techniques and expand datasets to incorporate broader pathological conditions, thus widening the clinical applicability and robustness of such advanced models.


